Multi-scale spiking network model of macaque visual cortex: Simulation data
This repository holds the simulation data presented in Schmidt M, Bakker R, Shen K, Bezgin B, Diesmann M & van Albada SJ (2018) A multi-scale layer-resolved spiking network model of resting-state dynamics in macaque cortex. PLOS Computational Biology (accepted).

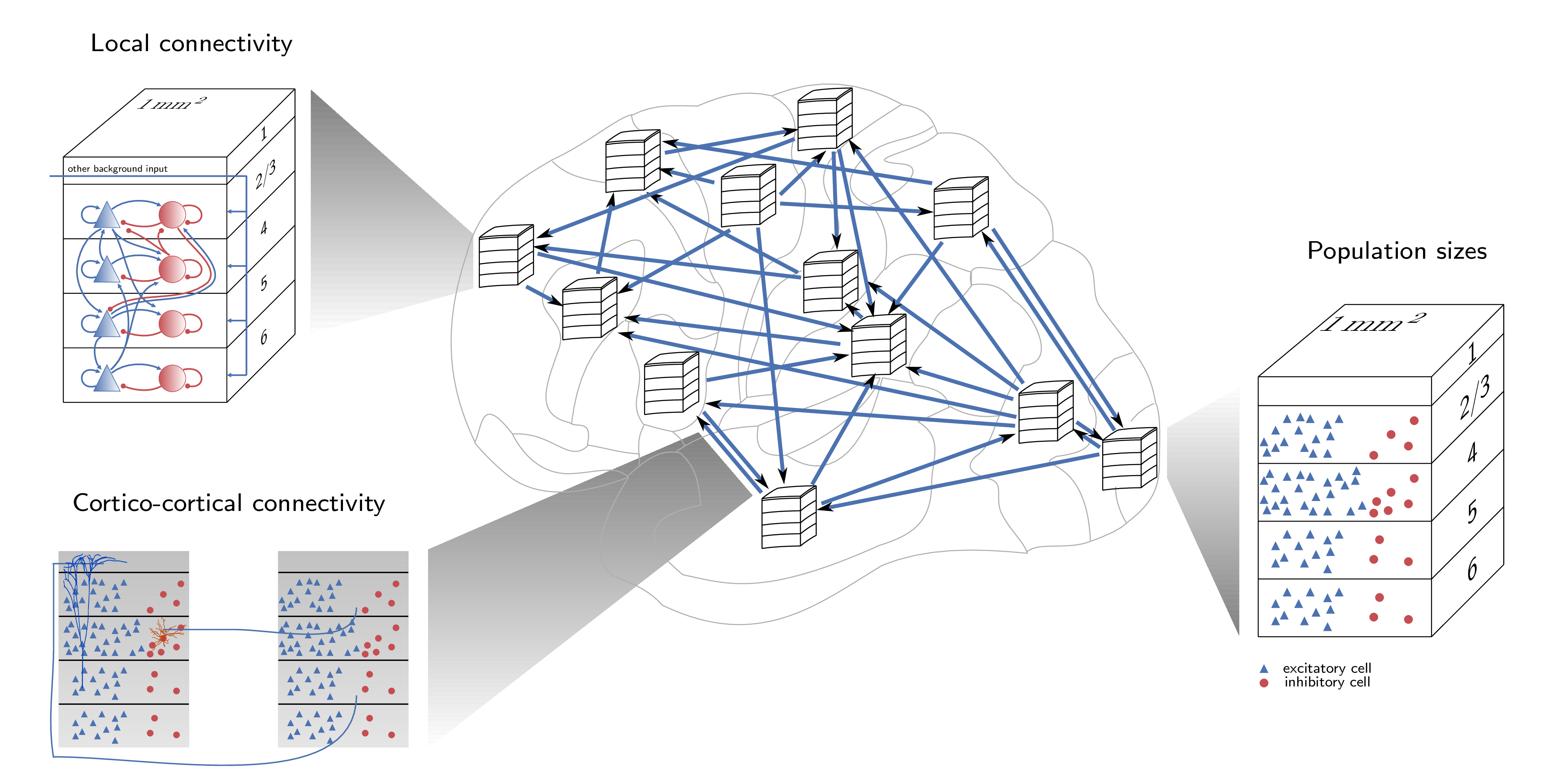
The simulation code of the model and the figure scripts for the paper reside in a public github repository available under the following url: https://github.com/INM-6/multi-area-model.
Simulations in this repository
This repository contains data from 17 different simulations that are presented in the paper. They are identified by their unique hashed labels that are generated from the parameter sets used in the respective simulation.
Here is a table that summarized the simulations and their most important parameters:
To reduce the required space, we here refrain from publishing the original spiking data (~150 GB) and publish only the post-processed data presented in the figures of Schmidt et al.
Citation
If you use these data, we ask you to cite our paper in your publication.
If you have questions regarding the code or scientific content, please create an issue on the simulation code repository on github: https://github.com/INM-6/multi-area-model.

