37 changed files with 1556 additions and 0 deletions
+ 11
- 0
README.md
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 57
- 0
balanced_network/Snakefile
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
balanced_network/images/correlations/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal_seed1_original.png

BIN
balanced_network/images/eigenspectrum/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal_original.png

BIN
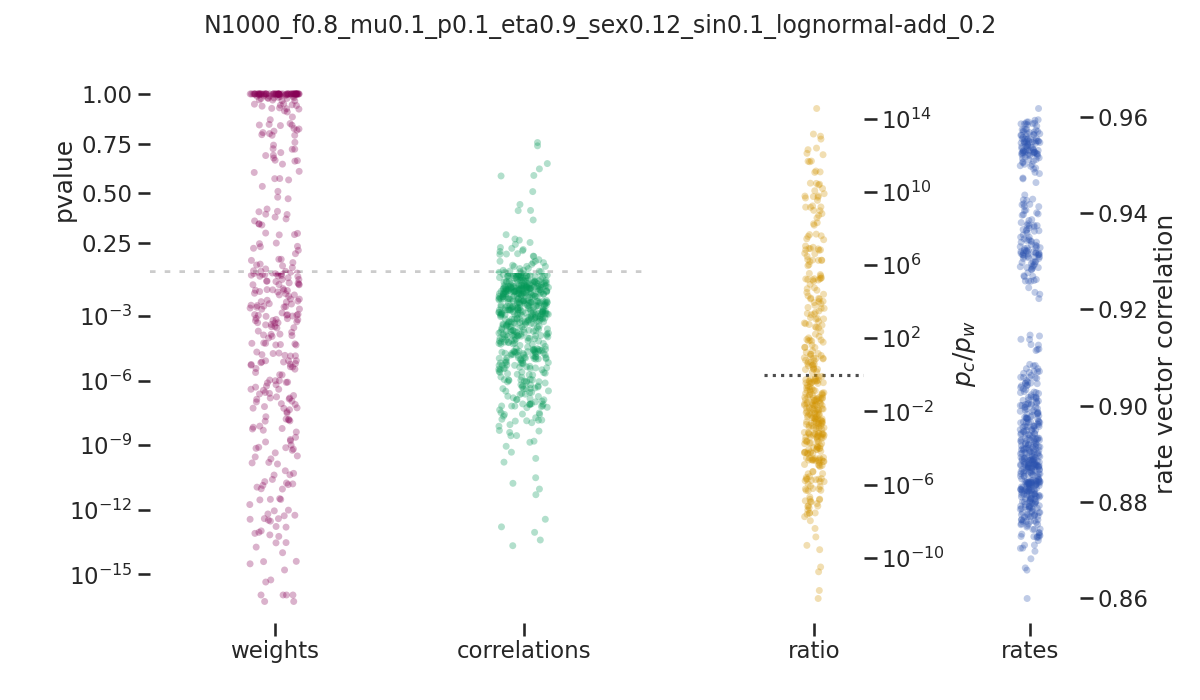
balanced_network/images/pvalue_overview/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-add_0.2.png
BIN
balanced_network/images/pvalue_overview/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-add_12800.png

BIN
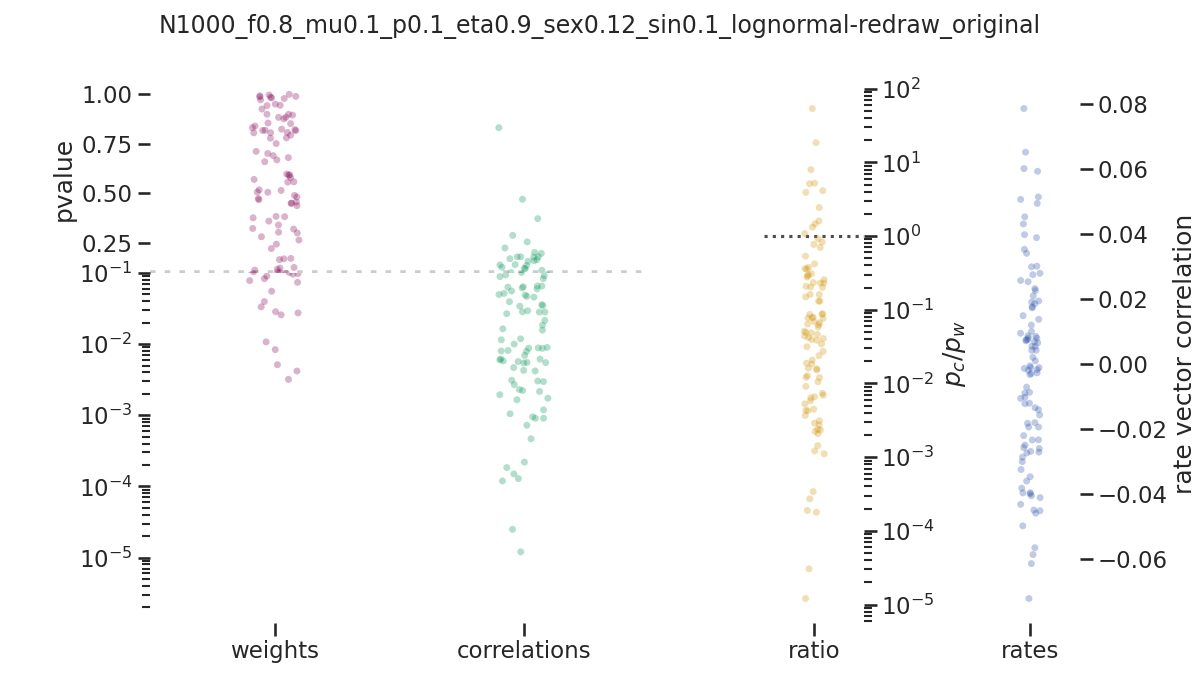
balanced_network/images/pvalue_overview/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-redraw_original.png
BIN
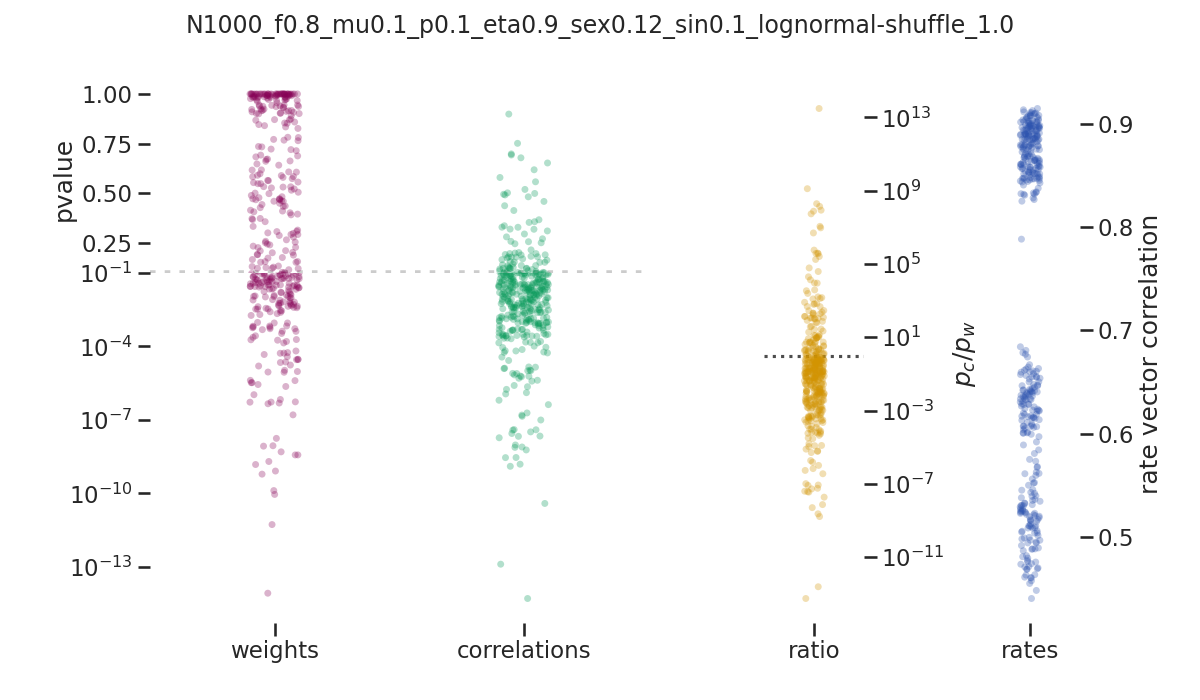
balanced_network/images/pvalue_overview/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-shuffle_1.0.png
BIN
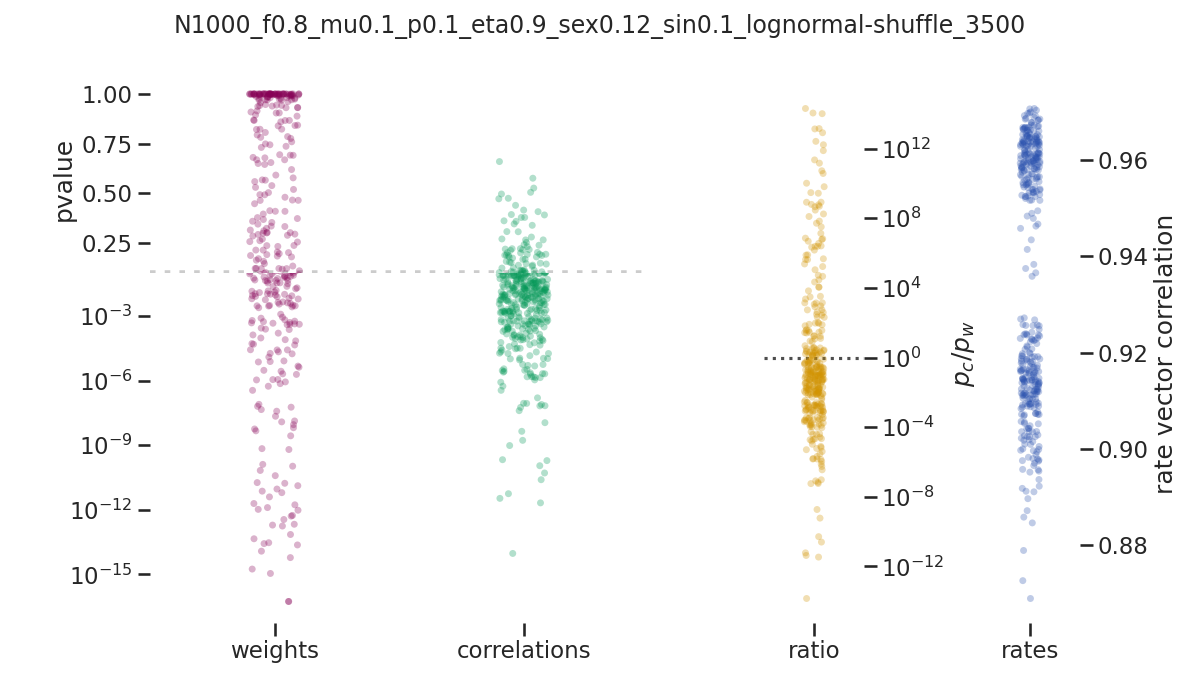
balanced_network/images/pvalue_overview/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-shuffle_3500.png
BIN
balanced_network/images/pvalue_trend/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-add_0.2.png
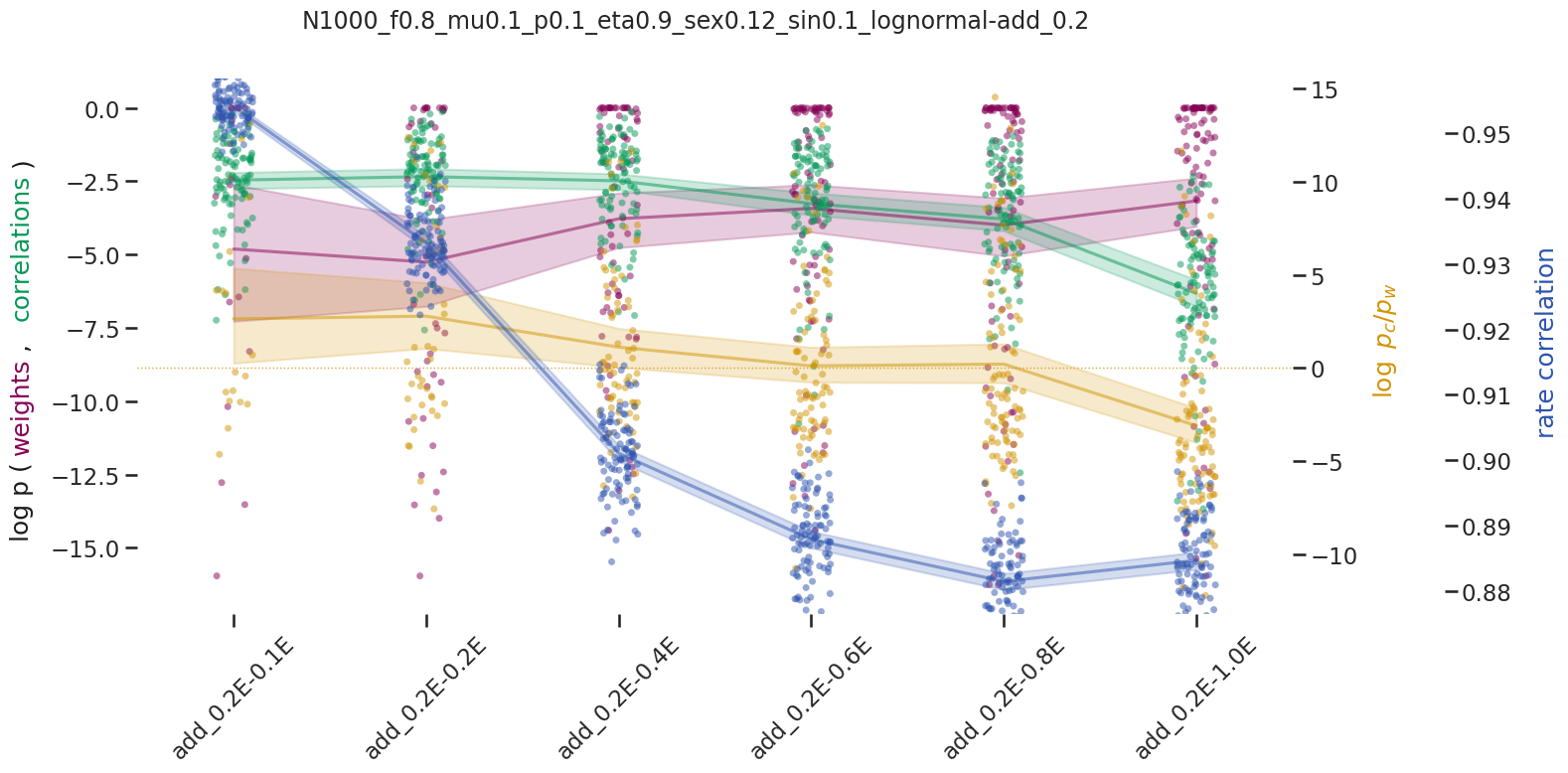
BIN
balanced_network/images/pvalue_trend/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-add_12800.png

BIN
balanced_network/images/pvalue_trend/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-shuffle_1.0.png
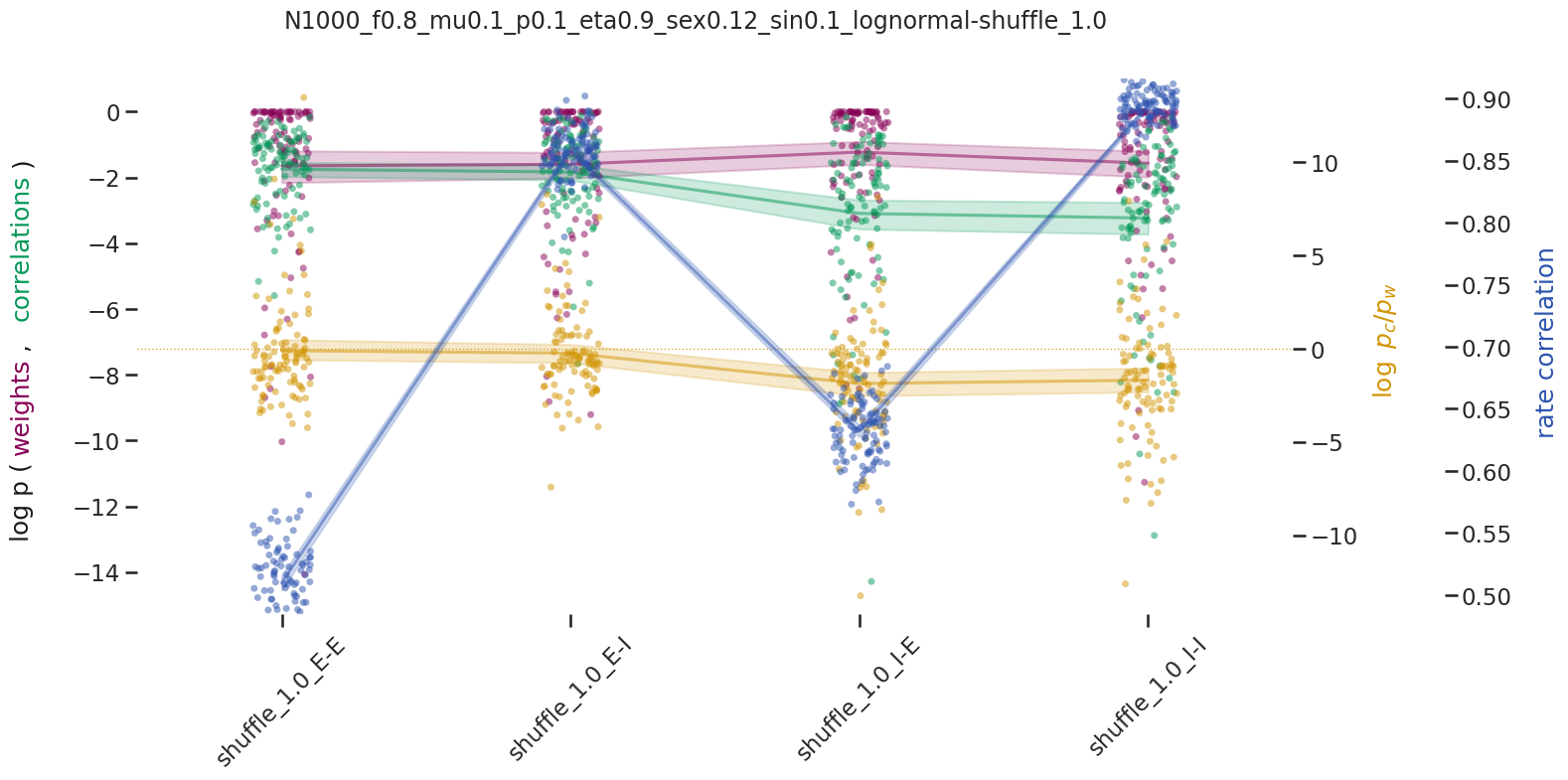
BIN
balanced_network/images/pvalue_trend/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal-shuffle_3500.png
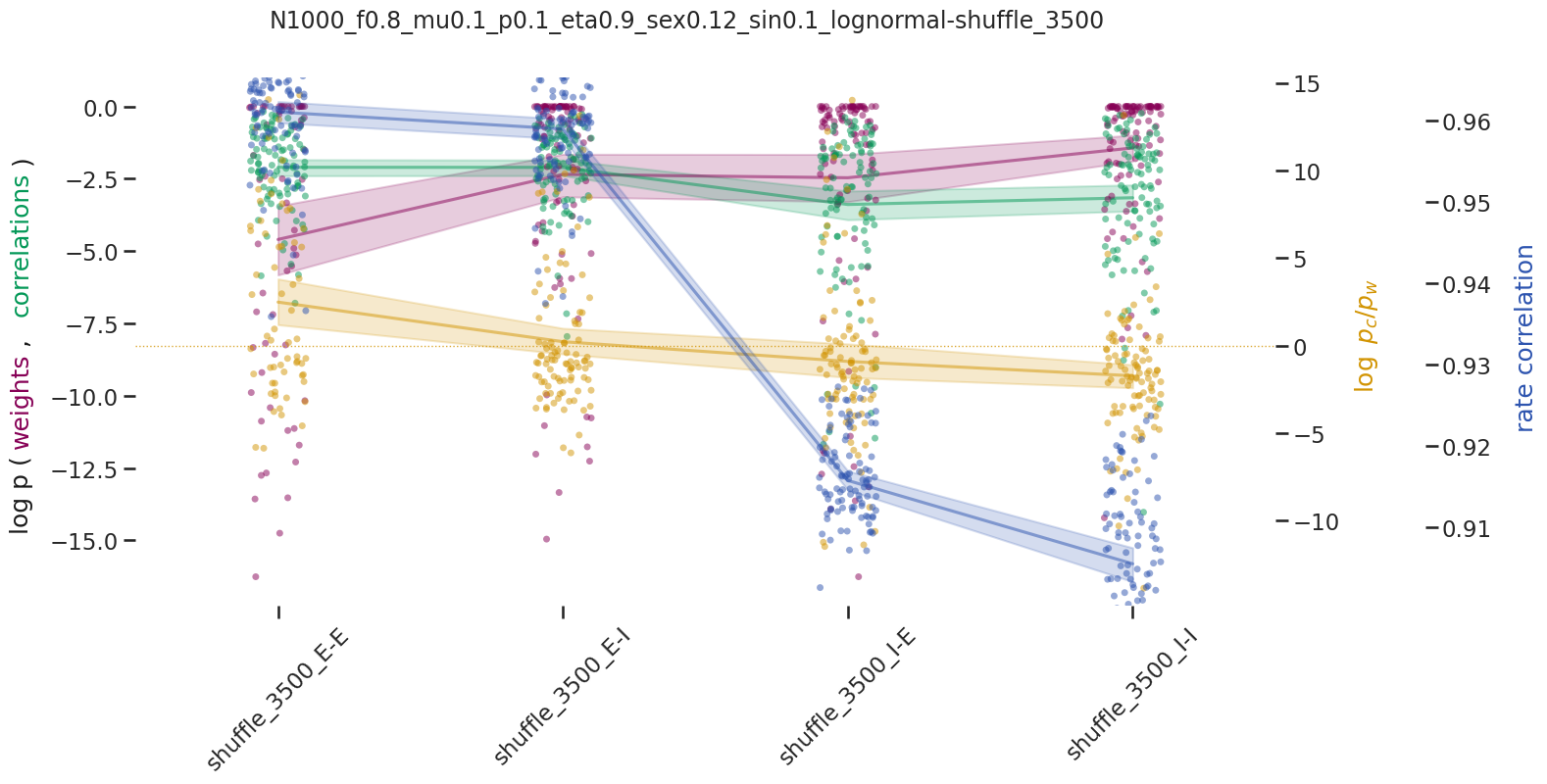
BIN
balanced_network/images/rasterplot/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal_seed1_original_spikes55000-60000ms.png

BIN
balanced_network/images/weights/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal_seed1_original.png

+ 14
- 0
balanced_network/scripts/plot_eigenspectrum.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 4
- 0
balanced_network/scripts/plot_pvalue_overview.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 4
- 0
balanced_network/scripts/plot_pvalue_trend.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
File diff suppressed because it is too large
+ 485
- 0
balanced_network/scripts/plot_rewiring.ipynb
File diff suppressed because it is too large
+ 120
- 0
eigenangle_basics.ipynb
+ 55
- 0
eigenangles.bib
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
paper_figures/CPP-pairwise_comparison.png
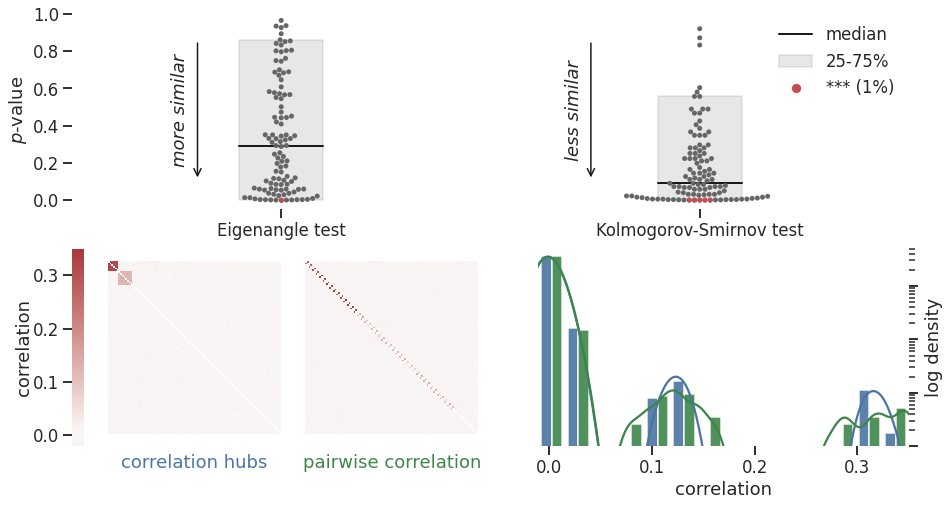
BIN
paper_figures/C_SpiNNaker_comparison.png

BIN
paper_figures/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal_original.png

BIN
paper_figures/N1000_f0.8_mu0.1_p0.1_eta0.9_sex0.12_sin0.1_lognormal_seed1_original_spikes55000-60000ms.png

BIN
paper_figures/figure_1/angle_distributions.png
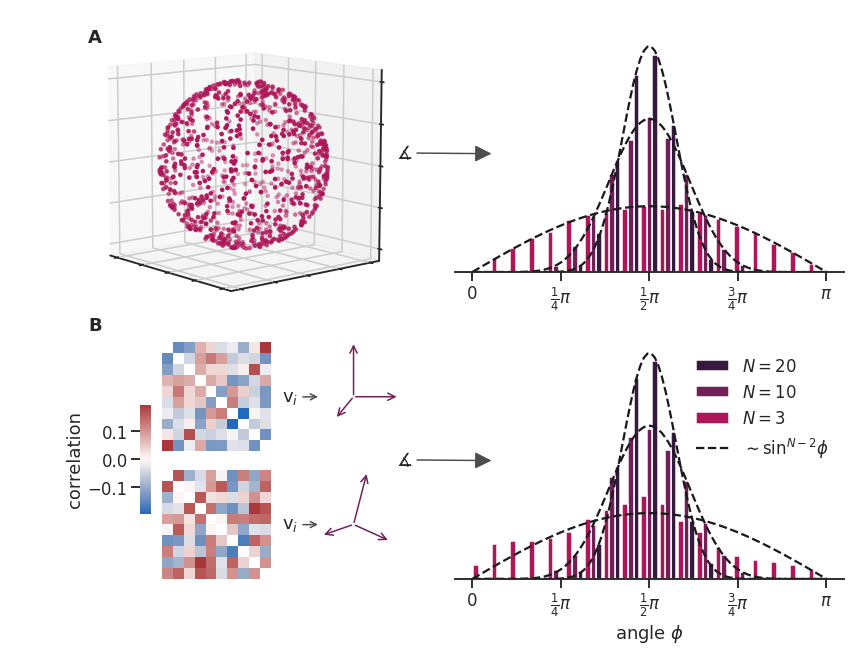
File diff suppressed because it is too large
+ 12
- 0
paper_figures/figure_1/figure_1.ipynb
File diff suppressed because it is too large
+ 24
- 0
paper_figures/figure_2/figure_2.ipynb
BIN
paper_figures/figure_2/test_construction.png

BIN
paper_figures/rewiring_comparisons_absolute_synapse_number_median.png

BIN
paper_figures/rewiring_comparisons_relative_synapse_number_median.png

BIN
paper_figures/stochastic_activity_similarity.png
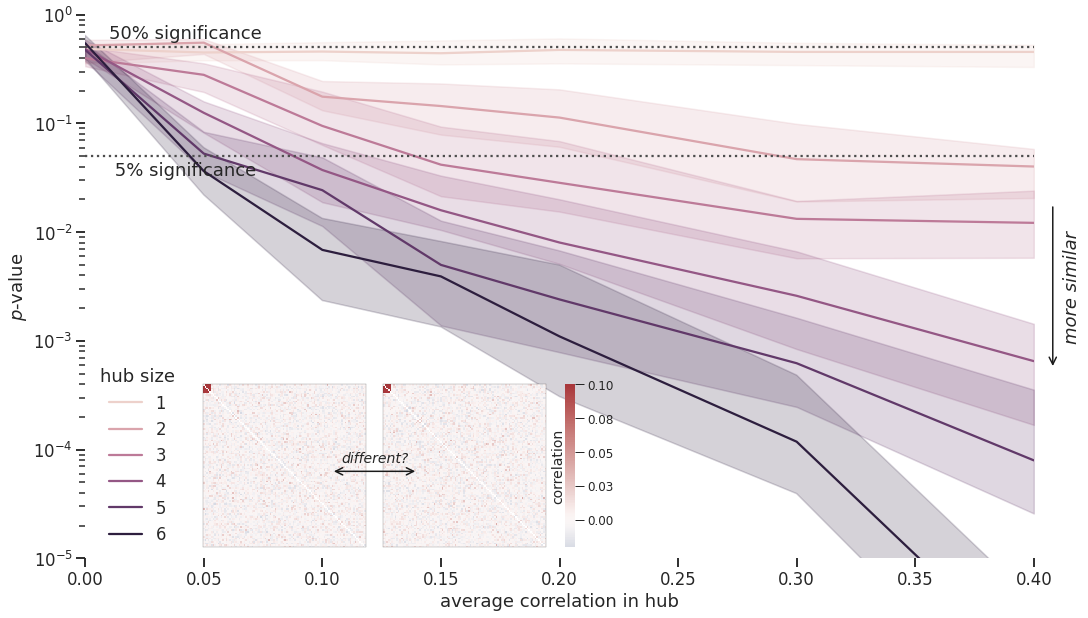
+ 4
- 0
polychrony_network/README.md
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 25
- 0
polychrony_network/scripts/plot_C-SpiNNaker_comparison.ipynb
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 4
- 0
scripts/plot_matrix.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||