README.md 5.7 KB
Working with model objects
Install
Download and install R https://www.r-project.org (and RStudio https://www.rstudio.com/products/rstudio/).
Go to R console or open scripts/README.Rmd in RStudio.
Install these packages (this needs to be done once).
if(!require(brms)){
install.packages(c("brms", "here"))
library(brms)
library(here)
}
## Loading required package: brms
## Loading required package: Rcpp
## Loading 'brms' package (version 2.14.4). Useful instructions
## can be found by typing help('brms'). A more detailed introduction
## to the package is available through vignette('brms_overview').
##
## Attaching package: 'brms'
## The following object is masked from 'package:stats':
##
## ar
Run
In R console OR in .Rmd file:
- Load packages to R environment.
library(brms)
library(here)
- Load the model object and print model summary.
m <- readRDS(here("models/anticons_detool.rds"))
print(m)
## Family: bernoulli
## Links: mu = logit
## Formula: anticons ~ de_tool
## Data: data (Number of observations: 2109)
## Samples: 4 chains, each with iter = 2000; warmup = 1000; thin = 1;
## total post-warmup samples = 4000
##
## Population-Level Effects:
## Estimate Est.Error l-95% CI u-95% CI Rhat Bulk_ESS Tail_ESS
## Intercept -3.51 0.23 -4.00 -3.09 1.00 942 1128
## de_tooldeseq 2.66 0.25 2.19 3.18 1.00 1080 1153
## de_tooledger 3.57 0.26 3.07 4.10 1.00 1051 1435
## de_toollimma 3.58 0.42 2.77 4.42 1.00 1693 2093
## de_toolunknown 2.46 0.25 1.99 2.98 1.00 1052 1153
##
## Samples were drawn using sampling(NUTS). For each parameter, Bulk_ESS
## and Tail_ESS are effective sample size measures, and Rhat is the potential
## scale reduction factor on split chains (at convergence, Rhat = 1).
- Get fitted coefficients with 95% credible intervals.
posterior_summary(m)
## Estimate Est.Error Q2.5 Q97.5
## b_Intercept -3.514582 0.2300317 -4.001753 -3.088126
## b_de_tooldeseq 2.656732 0.2492621 2.188271 3.183202
## b_de_tooledger 3.571890 0.2642955 3.074863 4.103799
## b_de_toollimma 3.579595 0.4163380 2.773039 4.424586
## b_de_toolunknown 2.461885 0.2476724 1.990004 2.975438
## lp__ -969.443536 1.6153116 -973.461001 -967.323674
- Get the full posterior.
post <- posterior_samples(m)
head(post)
## b_Intercept b_de_tooldeseq b_de_tooledger b_de_toollimma b_de_toolunknown lp__
## 1 -3.300792 2.355140 3.418613 3.433826 2.191998 -968.0241
## 2 -3.485360 2.606697 3.559653 3.530865 2.329601 -967.5360
## 3 -3.628822 2.705505 3.763745 3.932852 2.560540 -967.7857
## 4 -3.621123 2.917484 3.643992 3.647139 2.545874 -968.5439
## 5 -3.795040 2.929855 3.797475 3.520305 2.633857 -968.9733
## 6 -3.248306 2.532679 3.443725 3.528865 2.130235 -969.7214
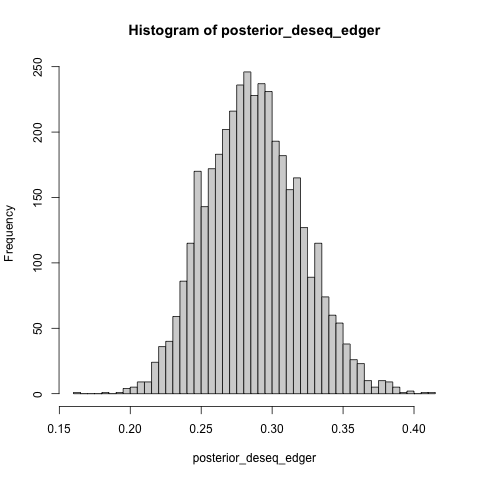
- What is the estimated difference in the proportion of anti-conservative p value histograms between DESeq2 and EdgeR?
posterior_deseq_edger <- inv_logit_scaled(post$b_de_tooldeseq - post$b_de_tooledger)
hist(posterior_deseq_edger, breaks = 40)
- The posterior summary for the effect size.
posterior_summary(posterior_deseq_edger)
## Estimate Est.Error Q2.5 Q97.5
## [1,] 0.2870903 0.03315653 0.2267546 0.3541821
The estimated effect size is somewhere between 22.7 and 35.4 percentage points.
- Get data from model object.
data <- m$data
head(data)
## anticons de_tool
## 1 1 unknown
## 2 0 unknown
## 3 1 edger
## 4 0 cuffdiff
## 5 0 limma
## 6 0 unknown
- Extract stan code from model object. This is the fullest model description.
stancode(m)
## // generated with brms 2.15.0
## functions {
## }
## data {
## int<lower=1> N; // total number of observations
## int Y[N]; // response variable
## int<lower=1> K; // number of population-level effects
## matrix[N, K] X; // population-level design matrix
## int prior_only; // should the likelihood be ignored?
## }
## transformed data {
## int Kc = K - 1;
## matrix[N, Kc] Xc; // centered version of X without an intercept
## vector[Kc] means_X; // column means of X before centering
## for (i in 2:K) {
## means_X[i - 1] = mean(X[, i]);
## Xc[, i - 1] = X[, i] - means_X[i - 1];
## }
## }
## parameters {
## vector[Kc] b; // population-level effects
## real Intercept; // temporary intercept for centered predictors
## }
## transformed parameters {
## }
## model {
## // likelihood including constants
## if (!prior_only) {
## target += bernoulli_logit_glm_lpmf(Y | Xc, Intercept, b);
## }
## // priors including constants
## target += student_t_lpdf(Intercept | 3, 0, 2.5);
## }
## generated quantities {
## // actual population-level intercept
## real b_Intercept = Intercept - dot_product(means_X, b);
## }
This document
This README.md was generated by running:
rmarkdown::render("scripts/README.Rmd", output_file = here::here("README.md"))