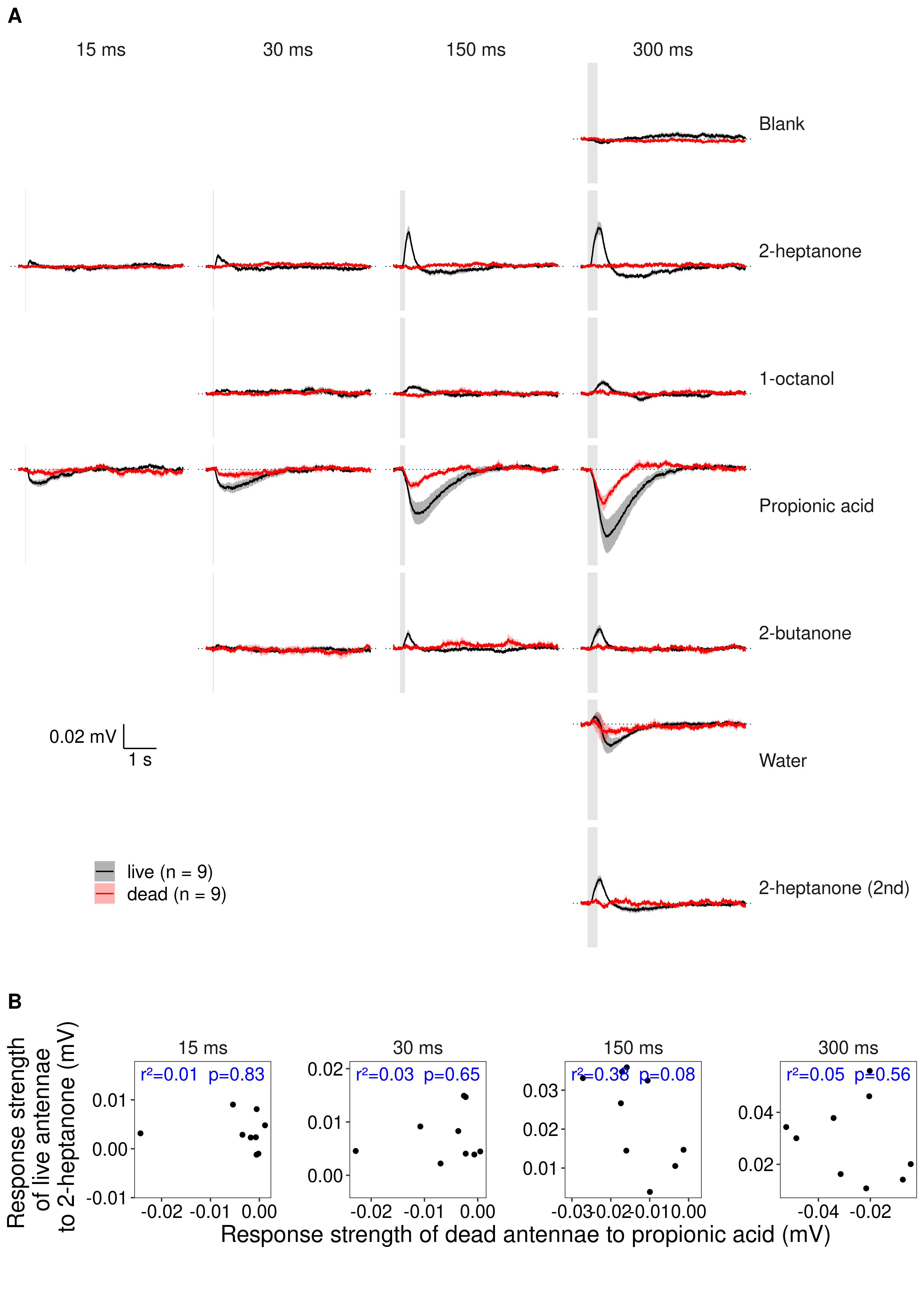
|
|
@@ -0,0 +1,388 @@
|
|
|
+##' Data Analysis of Electroantennogram Data from two different Stonefly Ecotypes
|
|
|
+##' (full-winged and wing-reduced)
|
|
|
+##'
|
|
|
+##' R routine to reproduce the Fig S2 from the manuscript entitled
|
|
|
+##' 'Rapid evolution of olfactory degradation in recently flightless
|
|
|
+##' insects'.
|
|
|
+##'
|
|
|
+##
|
|
|
+
|
|
|
+# load R package
|
|
|
+library(dplyr)
|
|
|
+library(tidyr)
|
|
|
+library(ggplot2)
|
|
|
+library(tibble)
|
|
|
+library(scales)
|
|
|
+
|
|
|
+
|
|
|
+# clean up environment
|
|
|
+rm(list=ls())
|
|
|
+
|
|
|
+
|
|
|
+# set working directory
|
|
|
+currentpath <- dirname(rstudioapi::getSourceEditorContext()$path)
|
|
|
+setwd(currentpath)
|
|
|
+getwd()
|
|
|
+
|
|
|
+
|
|
|
+# load function to calculate various response dynamic measures
|
|
|
+source('./functions/responseDynamics.R')
|
|
|
+source('./functions/utils.R')
|
|
|
+
|
|
|
+
|
|
|
+fig_path <- "../Figures"
|
|
|
+
|
|
|
+
|
|
|
+# aspect ratio (as y/x) for trace plots
|
|
|
+ar <- 2/3
|
|
|
+
|
|
|
+### colors for stoneflies ###
|
|
|
+fw_sm_l <- "#000000" ## black
|
|
|
+fw_sm_d <- "red"
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+data2 <- read.csv("../Datasets/Dataset2.csv", stringsAsFactors = FALSE)
|
|
|
+
|
|
|
+
|
|
|
+# automatically read in amplification from info file
|
|
|
+multiply_voltage <- 1/data2$Amplification
|
|
|
+
|
|
|
+## correct voltage values for the different amplification factor
|
|
|
+data2[, paste0("t", 1:5000)] <- data2[, paste0("t", 1:5000)] * multiply_voltage #in Volt
|
|
|
+data2[, paste0("t", 1:5000)] <- data2[, paste0("t", 1:5000)] * 1000 # in millivolt
|
|
|
+
|
|
|
+
|
|
|
+# convert ID to a factor
|
|
|
+data2$ID <- as.factor(data2$ID)
|
|
|
+data2$AntennaState <- as.factor(data2$AntennaState)
|
|
|
+data2 <- droplevels(data2)
|
|
|
+
|
|
|
+
|
|
|
+# reorder the factor levels of column Odor
|
|
|
+data2$Odor <- factor(data2$Odor, levels = c("Blank", "2Heptanone", "1Octanol", "PropionicAcid",
|
|
|
+ "2Butanone", "Water", "2HeptanoneTest"),
|
|
|
+ labels = c("Blank", "2-heptanone", "1-octanol", "Propionic acid",
|
|
|
+ "2-butanone", "Water", "2-heptanone (2nd)"))
|
|
|
+
|
|
|
+
|
|
|
+# data2$antpart[data2$Winged=="base"] <- "middle"
|
|
|
+data2$antpart[data2$Winged=="tip"] <- "distal"
|
|
|
+data2$antpart <- as.factor(data2$antpart)
|
|
|
+data2$AntennaState <- factor(data2$AntennaState,
|
|
|
+ levels = levels(data2$AntennaState),
|
|
|
+ labels = c("live", "dead"))
|
|
|
+data2 <- droplevels(data2)
|
|
|
+
|
|
|
+data2$NewGroup <- paste(data2$antpart, data2$AntennaState)
|
|
|
+data2$NewGroup <- as.factor(data2$NewGroup)
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+# create median filter mean traces (only for non-10Hz stimulations)
|
|
|
+med <- as.data.frame(t(apply(data2[data2$PulseDuration != 50, paste0("t", 1:5000)], 1, runmed, k=11)))
|
|
|
+data2[data2$PulseDuration != 50, paste0("t", 1:5000)] <- med
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+# add response characteristcs
|
|
|
+y1 <- as.data.frame(t(apply(data2[, paste0("t", 201:5000)], 1, responseDynamics)))
|
|
|
+
|
|
|
+colnames(y1) <- c("Timeto10Max",
|
|
|
+ "Timeto90Max",
|
|
|
+ "TimetoMax",
|
|
|
+ "TimeFromMaxto90",
|
|
|
+ "TimeFromMaxto10",
|
|
|
+ "Time10to90Max",
|
|
|
+ "Time90to10Max",
|
|
|
+ "Timeto10Min",
|
|
|
+ "Timeto90Min",
|
|
|
+ "TimetoMin",
|
|
|
+ "TimeFromMinto90",
|
|
|
+ "TimeFromMinto10",
|
|
|
+ "Time10to90Min",
|
|
|
+ "Time90to10Min",
|
|
|
+ "MeanMax",
|
|
|
+ "MeanMin")
|
|
|
+
|
|
|
+
|
|
|
+data2 <- cbind(as.data.frame(data2), y1)
|
|
|
+
|
|
|
+data2$max <- apply(data2[, paste0("t", 201:5000)], 1, max)
|
|
|
+data2$min <- apply(data2[, paste0("t", 201:5000)], 1, min)
|
|
|
+data2 <- data2 %>%
|
|
|
+ mutate(mima = ifelse(Odor == "Propionic acid" |
|
|
|
+ (Odor == "Water" & (abs(min) > max)), min, max))
|
|
|
+
|
|
|
+data2 <- data2 %>%
|
|
|
+ mutate(Mean = ifelse(Odor == "Propionic acid" |
|
|
|
+ (Odor == "Water" & (abs(min) > max)), MeanMin, MeanMax))
|
|
|
+
|
|
|
+
|
|
|
+#######################################
|
|
|
+
|
|
|
+## generate plot function for difference in factor NewGroup
|
|
|
+
|
|
|
+plot_Antenna <- function(data, group) {
|
|
|
+
|
|
|
+ d_mean <- data %>%
|
|
|
+ filter(Group==group) %>%
|
|
|
+ group_by(AntennaState, Odor, PulseDuration) %>%
|
|
|
+ summarise_at(.funs = mean, .vars=paste0("t", 1:5000))
|
|
|
+
|
|
|
+
|
|
|
+ # calculate the standard error over all odor-pulseduration configurations
|
|
|
+ d_se <- data %>%
|
|
|
+ filter(Group==group) %>%
|
|
|
+ group_by(AntennaState, Odor, PulseDuration) %>%
|
|
|
+ summarise_at(.funs = se, .vars=paste0("t", 1:5000))
|
|
|
+
|
|
|
+ # calculate sample size (n) over all odor-pulseduration configurations
|
|
|
+ d_n <- data %>%
|
|
|
+ filter(Group==group) %>%
|
|
|
+ group_by(AntennaState, Odor, PulseDuration) %>%
|
|
|
+ summarise(N=n()) %>%
|
|
|
+ group_by(AntennaState) %>%
|
|
|
+ summarise(N=unique(N))
|
|
|
+ d_n <- unite(d_n, col = "AntennaState", sep = " (n = ")
|
|
|
+ d_n$t <- ")"
|
|
|
+ d_n <- unite(d_n, col = "AntennaState", sep = "")
|
|
|
+
|
|
|
+
|
|
|
+ d_all <- d_mean
|
|
|
+ data_se <- gather(d_se, time, se, t1:t5000, factor_key=TRUE)
|
|
|
+
|
|
|
+
|
|
|
+ d_all$PulseDuration <- factor(d_all$PulseDuration, levels = c(15, 30, 150, 300))
|
|
|
+ d_all$PulseDur <- factor(d_all$PulseDuration, levels = c(15, 30, 150, 300),
|
|
|
+ labels = c("15 ms", "30 ms", "150 ms", "300 ms"))
|
|
|
+
|
|
|
+
|
|
|
+ data_long <- gather(d_all, time, Voltage, t1:t5000, factor_key=TRUE)
|
|
|
+ data_long$time <- (as.numeric(gsub("t", "", data_long$time)) - 200)/1000
|
|
|
+ data_long$SE <- data_se$se
|
|
|
+ data_long$AntennaState <- droplevels(data_long$AntennaState)
|
|
|
+ data_long$AntennaState <- factor(data_long$AntennaState,
|
|
|
+ labels = t(d_n))
|
|
|
+
|
|
|
+ data_long$high <- data_long$Voltage + data_long$SE
|
|
|
+ data_long$low <- data_long$Voltage - data_long$SE
|
|
|
+
|
|
|
+
|
|
|
+ ## generate dummy values for a the same scale of y, but different ranges (max and min)
|
|
|
+ ranges <- data.frame("NegResp" = range(data_long[data_long$Odor=="Water" |
|
|
|
+ data_long$Odor=="Propionic acid", c("high", "low")]),
|
|
|
+ "PosResp" = range(data_long[data_long$Odor!="Water" &
|
|
|
+ data_long$Odor!="Propionic acid", c("high", "low")]),
|
|
|
+ "Range"= c("min", "max"))
|
|
|
+
|
|
|
+ roundUp <- function(x, to) {
|
|
|
+ to*(x%/%to + as.logical(x%%to))
|
|
|
+ }
|
|
|
+ max_diff <- roundUp(max(diff(ranges$NegResp), diff(ranges$PosResp)), to = 0.01)
|
|
|
+
|
|
|
+
|
|
|
+ max_diff/2
|
|
|
+ ranges$NegResp_new <- c((ranges$NegResp[1] + (diff(ranges$NegResp)/2)) - max_diff/2,
|
|
|
+ (ranges$NegResp[1] + (diff(ranges$NegResp)/2)) + max_diff/2)
|
|
|
+ ranges$PosResp_new <- c((ranges$PosResp[1] + (diff(ranges$PosResp)/2)) - max_diff/2,
|
|
|
+ (ranges$PosResp[1] + (diff(ranges$PosResp)/2)) + max_diff/2)
|
|
|
+
|
|
|
+ df <- as.data.frame(table(data_long$Odor, data_long$PulseDuration))
|
|
|
+ df <- df[df$Freq!=0,]
|
|
|
+ colnames(df) <- c("Odor", "PulseDuration", "Freq")
|
|
|
+ df$range <- "min"
|
|
|
+ df$time <- 1
|
|
|
+
|
|
|
+ df2 <- df
|
|
|
+ df2$range <- "max"
|
|
|
+
|
|
|
+ df <- rbind(df, df2)
|
|
|
+
|
|
|
+ df$Voltage[(df$Odor=="Water" | df$Odor=="Propionic acid") & df$range=="min"] <- ranges$NegResp_new[1]
|
|
|
+ df$Voltage[(df$Odor=="Water" | df$Odor=="Propionic acid") & df$range=="max"] <- ranges$NegResp_new[2]
|
|
|
+ df$Voltage[(df$Odor!="Water" & df$Odor!="Propionic acid") & df$range=="min"] <- ranges$PosResp_new[1]
|
|
|
+ df$Voltage[(df$Odor!="Water" & df$Odor!="Propionic acid") & df$range=="max"] <- ranges$PosResp_new[2]
|
|
|
+
|
|
|
+ diff(range(df[df$Odor=="Water", "Voltage"]))
|
|
|
+
|
|
|
+ mids <- df %>%
|
|
|
+ filter(PulseDuration=="300") %>%
|
|
|
+ group_by(Odor) %>%
|
|
|
+ summarise(mid=Voltage[range=="max"] - ((Voltage[range=="max"] - Voltage[range=="min"])/2))
|
|
|
+
|
|
|
+
|
|
|
+ data_segm1 <- data.frame(y=-0.02, yend=c(0), x=3, xend=3,
|
|
|
+ PulseDuration=15,
|
|
|
+ Odor=c("Water"))
|
|
|
+ label1 <- data.frame(x=3, y=c(-0.01),
|
|
|
+ text=c("0.02 mV"),
|
|
|
+ PulseDuration= 15,
|
|
|
+ Odor=c("Water"))
|
|
|
+ data_segm2 <- data.frame(x=3, xend=4, y=-0.02, yend=-0.02,
|
|
|
+ PulseDuration= 15,
|
|
|
+ Odor=c("Water"))
|
|
|
+ label2 <- data.frame(x=3.5, y=-0.02,
|
|
|
+ text=c("1 s"),
|
|
|
+ PulseDuration= 15,
|
|
|
+ Odor="Water")
|
|
|
+
|
|
|
+
|
|
|
+ label3 <- data.frame(x=5, y=mids$mid,
|
|
|
+ text=levels(data_long$Odor),
|
|
|
+ PulseDuration=300,
|
|
|
+ Odor=levels(data_long$Odor))
|
|
|
+
|
|
|
+ col <- c(fw_sm_l, fw_sm_d)
|
|
|
+
|
|
|
+ df_sub <- df[df$range=="min",]
|
|
|
+ df_sub <- droplevels(df_sub)
|
|
|
+ df_sub$PulseDuration <- factor(df_sub$PulseDuration, levels = c(15, 30, 150, 300))
|
|
|
+
|
|
|
+ ggplot(data_long, aes(x=time, y=Voltage)) +
|
|
|
+ scale_color_manual(aesthetics = c("colour", "fill"), values = col) +
|
|
|
+ facet_grid(Odor ~ factor(PulseDuration,
|
|
|
+ levels = c(15, 30, 150, 300),
|
|
|
+ labels = c("15 ms", "30 ms", "150 ms", "300 ms")),
|
|
|
+ scales = "free_y") +
|
|
|
+ geom_rect(mapping=aes(xmin=0, xmax=as.numeric(as.character(PulseDuration))/1000,
|
|
|
+ ymin=-Inf, ymax=Inf),
|
|
|
+ fill="grey90", alpha=1) +
|
|
|
+ geom_hline(mapping = aes(yintercept = 0), color="black", size=0.3, linetype="dotted") +
|
|
|
+ geom_ribbon(aes(ymin=Voltage + SE, ymax=Voltage - SE, fill=AntennaState), alpha=0.3) +
|
|
|
+ geom_line(aes(colour = AntennaState)) +
|
|
|
+ theme_bw() +
|
|
|
+ geom_blank(df, mapping = aes(x=time, y=Voltage)) +
|
|
|
+ labs(x = "Time (s)", y="Voltage (mV)") +
|
|
|
+ theme(panel.grid.major = element_blank(),
|
|
|
+ panel.grid.minor = element_blank(),
|
|
|
+ axis.line = element_blank(),
|
|
|
+ axis.title = element_blank(),
|
|
|
+ axis.ticks = element_blank(),
|
|
|
+ aspect.ratio = ar,
|
|
|
+ plot.margin = unit(c(5.5, 5.5, 5.5, 5.5), "points"),
|
|
|
+ strip.background = element_rect(colour="NA", fill="NA"),
|
|
|
+ panel.border = element_blank(),
|
|
|
+ text = element_text(size=18),
|
|
|
+ axis.text=element_blank(),
|
|
|
+ strip.text.y = element_text(angle = 0, hjust = 0),
|
|
|
+ legend.position = c(0.2, 0.08),
|
|
|
+ legend.title = element_blank()) +
|
|
|
+ geom_segment(data=data_segm1,
|
|
|
+ aes(x=x, y=y, yend=yend, xend=xend)) +
|
|
|
+ geom_text(data = label1, aes(x=x, y=y, label=text), size=5, hjust=1.1) +
|
|
|
+ geom_segment(data=data_segm2,
|
|
|
+ aes(x=x, y=y, yend=yend, xend=xend)) +
|
|
|
+ geom_text(data = label2, aes(x=x, y=y, label=text), size=5, vjust=1.3) +
|
|
|
+ coord_cartesian(expand = TRUE,
|
|
|
+ clip = 'off')
|
|
|
+}
|
|
|
+
|
|
|
+
|
|
|
+## plot real data
|
|
|
+p1 <- plot_Antenna(data = data2,
|
|
|
+ group = "FenestrateZombie"); p1
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+subdat <- data2 %>%
|
|
|
+ dplyr::select(Mean, Odor, AntennaState, ID_new, PulseDuration) %>%
|
|
|
+ group_by(AntennaState, ID_new, PulseDuration) %>%
|
|
|
+ pivot_wider(names_from = Odor, values_from = Mean)
|
|
|
+
|
|
|
+colnames(subdat) <- c("AntennaState","ID_new","PulseDuration","Blank","HPT",
|
|
|
+ "OCT","PropionicAcid","BTN","Water","HPT2")
|
|
|
+
|
|
|
+subdat2 <- subdat %>%
|
|
|
+ group_by(ID_new, PulseDuration) %>%
|
|
|
+ summarise("HeptAlive"=HPT[AntennaState=="live"], "PropDead"=PropionicAcid[AntennaState=="dead"])
|
|
|
+
|
|
|
+subdat2$PulseDuration <- factor(subdat2$PulseDuration,
|
|
|
+ levels = c(15, 30, 150, 300))
|
|
|
+
|
|
|
+subdat2$pvals <- NA
|
|
|
+
|
|
|
+## Create a list holding the p-values for regressions on each PulseDuration
|
|
|
+subdat2 <- subdat2 %>%
|
|
|
+ group_by(PulseDuration) %>%
|
|
|
+ mutate(HeptAlive_sc=scale(HeptAlive), PropDead_sc=scale(PropDead))
|
|
|
+
|
|
|
+subdat2$ID_new <- as.factor(subdat2$ID_new)
|
|
|
+
|
|
|
+NEWDAT <- as.data.frame(matrix(NA, ncol = 7, nrow = 4))
|
|
|
+colnames(NEWDAT) <- c("PulseDuration", "pval", "r2", "xmin", "xmax", "ymin", "ymax")
|
|
|
+
|
|
|
+NEWDAT$PulseDuration <- as.character(unique(subdat2$PulseDuration))
|
|
|
+
|
|
|
+par(mfrow=c(1,4))
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+for (i in as.character(unique(subdat2$PulseDuration))) {
|
|
|
+ dat <- subdat2[subdat2$PulseDuration==i, ]
|
|
|
+
|
|
|
+ mod <- lm(HeptAlive ~ PropDead, data = dat)
|
|
|
+
|
|
|
+ ry <- range(dat$HeptAlive)
|
|
|
+ rx <- range(dat$PropDead)
|
|
|
+ rxy <- max(abs(diff(ry)), abs(diff(rx)))
|
|
|
+ xlim <- c((rx[1] + abs(diff(rx))/2) - rxy/2, (rx[1] + abs(diff(rx))/2) + rxy/2)
|
|
|
+ ylim <- c((ry[1] + abs(diff(ry))/2) - rxy/2, (ry[1] + abs(diff(ry))/2) + rxy/2)
|
|
|
+
|
|
|
+ plot(HeptAlive ~ PropDead, data = dat, pch=21, las=1,
|
|
|
+ cex.lab=1.2, xlim=xlim,
|
|
|
+ ylim=ylim, main=i)
|
|
|
+ abline(mod)
|
|
|
+ summary(mod)
|
|
|
+ m <- summary(mod)
|
|
|
+ print(m)
|
|
|
+
|
|
|
+ NEWDAT[NEWDAT$PulseDuration==i, "pval"] <- round(m$coefficients[2, 4], digits = 2)
|
|
|
+ NEWDAT[NEWDAT$PulseDuration==i, "r2"] <- round(m$r.squared, digits = 2)
|
|
|
+ NEWDAT[NEWDAT$PulseDuration==i, c("xmin", "xmax")] <- xlim
|
|
|
+ NEWDAT[NEWDAT$PulseDuration==i, c("ymin", "ymax")] <- ylim
|
|
|
+}
|
|
|
+
|
|
|
+
|
|
|
+subdat2$PulseDuration <- factor(subdat2$PulseDuration,
|
|
|
+ c(15, 30, 150, 300),
|
|
|
+ c("15 ms", "30 ms", "150 ms", "300 ms"))
|
|
|
+
|
|
|
+NEWDAT$PulseDuration <- factor(NEWDAT$PulseDuration,
|
|
|
+ c(15, 30, 150, 300),
|
|
|
+ c("15 ms", "30 ms", "150 ms", "300 ms"))
|
|
|
+
|
|
|
+dependencePlot <- ggplot(subdat2, aes(PropDead, HeptAlive)) +
|
|
|
+ geom_point() +
|
|
|
+ scale_x_continuous(breaks = pretty_breaks(n = 3)) +
|
|
|
+ scale_y_continuous(breaks = pretty_breaks(n = 3)) +
|
|
|
+ geom_blank(data=NEWDAT, mapping=aes(x=xmin, y=ymin)) +
|
|
|
+ geom_blank(data=NEWDAT, mapping=aes(x=xmax, y=ymax)) +
|
|
|
+ facet_wrap( . ~ PulseDuration, scales = "free", ncol=4) +
|
|
|
+ xlab("Response strength of dead antennae to propionic acid (mV)") +
|
|
|
+ ylab("Response strength \n of live antennae \n to 2-heptanone (mV)") +
|
|
|
+ theme_bw() +
|
|
|
+ theme(panel.grid.major = element_blank(),
|
|
|
+ panel.grid.minor = element_blank(),
|
|
|
+ panel.spacing = unit(2, "lines"),
|
|
|
+ plot.margin = unit(c(5.5, 5.5, 5.5, 5.5), "points"),
|
|
|
+ strip.background = element_rect(colour="NA", fill="NA"),
|
|
|
+ text = element_text(size=18),
|
|
|
+ aspect.ratio=1,
|
|
|
+ axis.text=element_text(colour="black")) +
|
|
|
+ geom_text(NEWDAT, mapping = aes(x=xmax, y=ymax, label=paste0("p=",pval)), size=5,
|
|
|
+ color="blue", hjust=1, vjust=1) +
|
|
|
+ geom_text(NEWDAT, mapping = aes(x=xmin, y=ymax, label=paste0("r²=", r2)), size=5,
|
|
|
+ color="blue", hjust=0, vjust=1)
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+FigS2 <- cowplot::plot_grid(p1, dependencePlot,
|
|
|
+ labels = c("A", "B"), label_size = 16,
|
|
|
+ ncol = 1, nrow = 2, rel_heights = c(3.2,1))
|
|
|
+
|
|
|
+FigS2
|
|
|
+
|
|
|
+ggsave("FigureS2.pdf", path = fig_path, width = 10, height = 14)
|
|
|
+ggsave("FigureS2.jpg", path = fig_path, width = 10, height = 14)
|