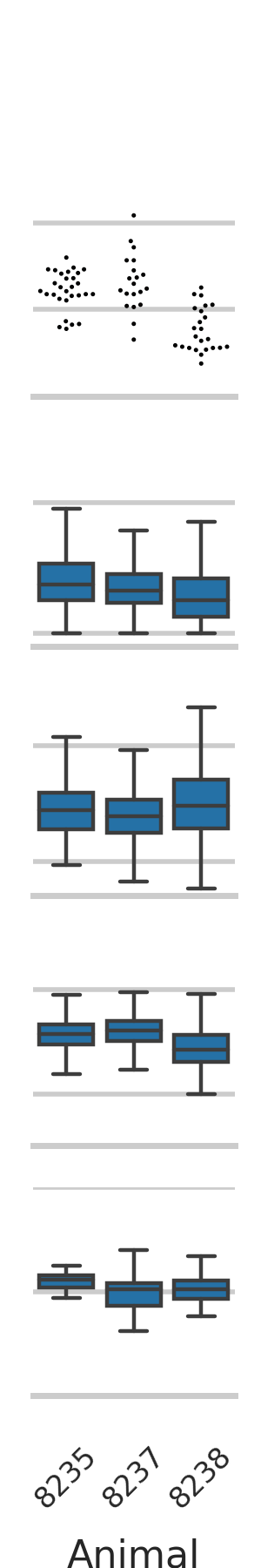
23 changed files with 6239 additions and 7029 deletions
+ 6
- 4
validation/calcium_imaging/Figure2_super_compact.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
validation/calcium_imaging/Validation_motion.png
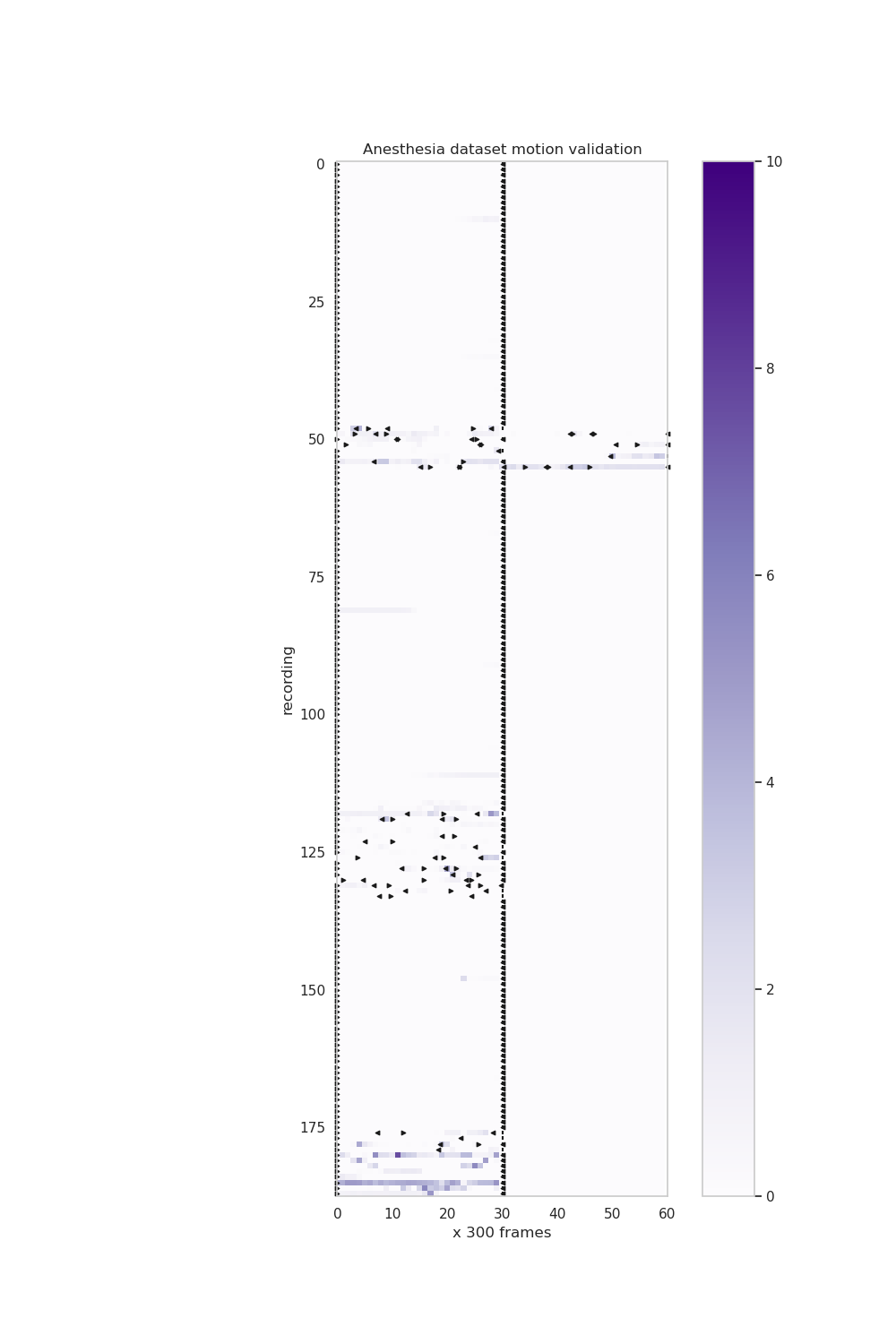
+ 35
- 25
validation/calcium_imaging/Validation_motion.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
File diff suppressed because it is too large
+ 2790
- 1912
validation/calcium_imaging/Validation_motion.svg
BIN
validation/calcium_imaging/Validation_stability_check_figure2_super_compact.png

File diff suppressed because it is too large
+ 780
- 827
validation/calcium_imaging/Validation_stability_check_figure2_super_compact.svg
BIN
validation/calcium_imaging/__pycache__/capipeline.cpython-38.pyc
+ 7
- 6
validation/calcium_imaging_transition_state/Figure2_super_compact.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
validation/calcium_imaging_transition_state/Validation_motion.png
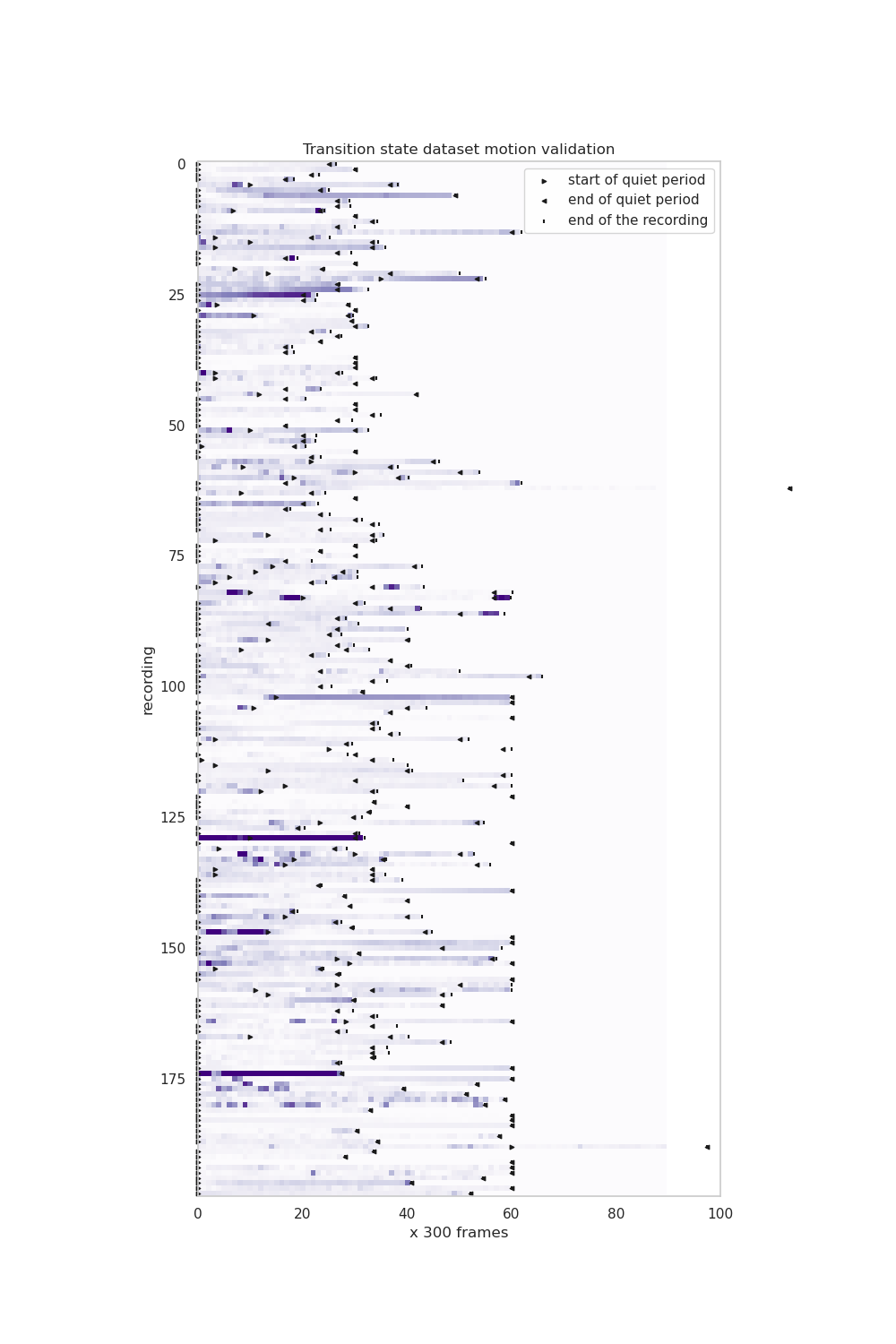
+ 10
- 13
validation/calcium_imaging_transition_state/Validation_motion.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
File diff suppressed because it is too large
+ 668
- 668
validation/calcium_imaging_transition_state/Validation_motion.svg
BIN
validation/calcium_imaging_transition_state/Validation_stability_check_figure2_super_compact.png
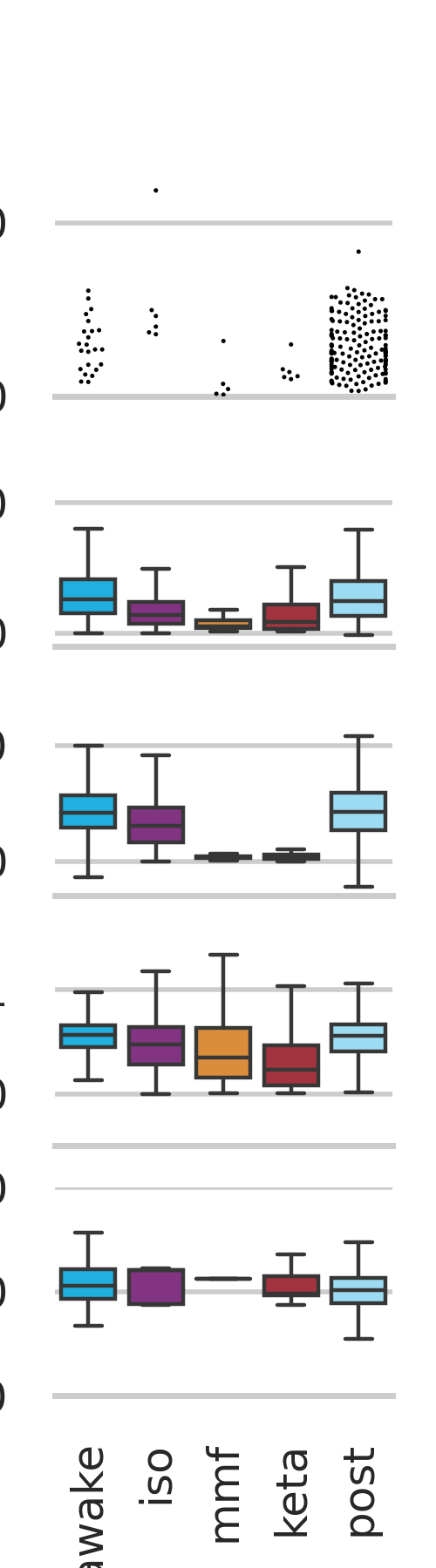
File diff suppressed because it is too large
+ 982
- 1055
validation/calcium_imaging_transition_state/Validation_stability_check_figure2_super_compact.svg
BIN
validation/calcium_imaging_transition_state/__pycache__/capipeline.cpython-38.pyc
+ 7
- 6
validation/sleep_data_calcium_imaging/Figure2_compact.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
validation/sleep_data_calcium_imaging/Validation_motion.png
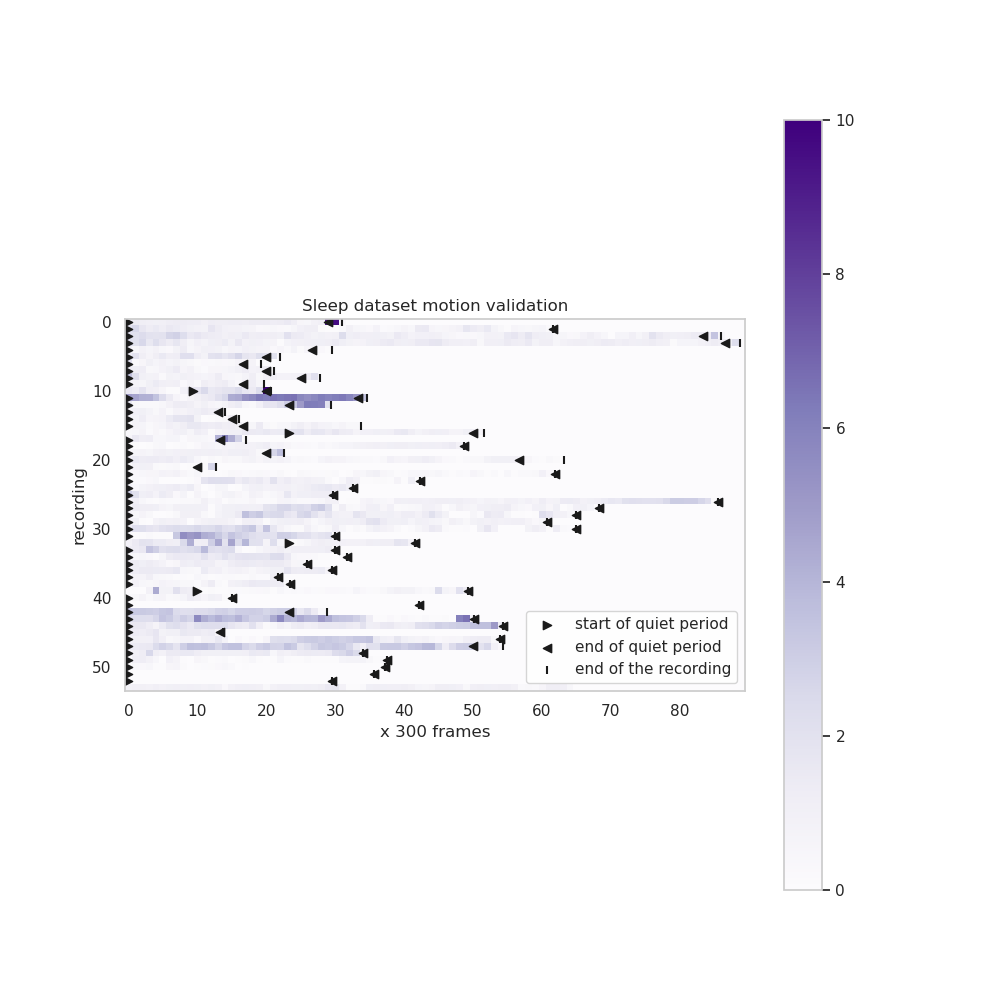
+ 12
- 7
validation/sleep_data_calcium_imaging/Validation_motion.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
File diff suppressed because it is too large
+ 344
- 304
validation/sleep_data_calcium_imaging/Validation_motion.svg
BIN
validation/sleep_data_calcium_imaging/Validation_stability_check_figure2.png
File diff suppressed because it is too large
+ 0
- 1735
validation/sleep_data_calcium_imaging/Validation_stability_check_figure2.svg
BIN
validation/sleep_data_calcium_imaging/Validation_stability_check_sleep_figure2.png