|
|
@@ -0,0 +1,216 @@
|
|
|
+#!/usr/bin/env python
|
|
|
+# coding: utf-8
|
|
|
+
|
|
|
+# ### Link to the file with meta information on recordings
|
|
|
+
|
|
|
+# In[14]:
|
|
|
+
|
|
|
+#import matplotlib.pyplot as plt
|
|
|
+#plt.rcParams["figure.figsize"] = (20,3)
|
|
|
+
|
|
|
+
|
|
|
+database_path = '/media/andrey/My Passport/GIN/Anesthesia_CA1/meta_data/meta_recordings_transition_state.xlsx'
|
|
|
+
|
|
|
+
|
|
|
+# ### Select the range of recordings for the analysis (see "Number" row in the meta data file)
|
|
|
+
|
|
|
+# In[4]:
|
|
|
+
|
|
|
+
|
|
|
+#rec = [x for x in range(0,198+1)]
|
|
|
+rec = [127,128]
|
|
|
+
|
|
|
+# In[1]:
|
|
|
+
|
|
|
+
|
|
|
+import numpy as np
|
|
|
+
|
|
|
+import numpy.ma as ma
|
|
|
+
|
|
|
+import matplotlib.pyplot as plt
|
|
|
+import matplotlib.ticker as ticker
|
|
|
+
|
|
|
+import pandas as pd
|
|
|
+import seaborn as sns
|
|
|
+
|
|
|
+import pickle
|
|
|
+import os
|
|
|
+
|
|
|
+sns.set()
|
|
|
+sns.set_style("whitegrid")
|
|
|
+
|
|
|
+from scipy.signal import medfilt
|
|
|
+
|
|
|
+from scipy.stats import skew, kurtosis, zscore
|
|
|
+
|
|
|
+from scipy import signal
|
|
|
+
|
|
|
+from sklearn.linear_model import LinearRegression, TheilSenRegressor
|
|
|
+
|
|
|
+plt.rcParams['figure.figsize'] = [16, 8]
|
|
|
+
|
|
|
+color_awake = (0,191/255,255/255)
|
|
|
+color_mmf = (245/255,143/255,32/255)
|
|
|
+color_keta = (181./255,34./255,48./255)
|
|
|
+color_iso = (143./255,39./255,143./255)
|
|
|
+
|
|
|
+color_post = (142/255,226/255,255/255)
|
|
|
+
|
|
|
+custom_palette ={'keta':color_keta, 'iso':color_iso,'mmf':color_mmf,'awake':color_awake,'post':color_post}
|
|
|
+
|
|
|
+
|
|
|
+# In[2]:
|
|
|
+
|
|
|
+
|
|
|
+from capipeline import *
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+df_estimators = pd.read_pickle("./transition_state_calcium_imaging_stability_validation.pkl")
|
|
|
+df_estimators['neuronID'] = df_estimators.index
|
|
|
+
|
|
|
+#df_estimators["animal"] = df_estimators["animal"]
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+print(np.unique(df_estimators["condition"]))
|
|
|
+
|
|
|
+df_estimators["CONDITION"] = df_estimators["condition"]
|
|
|
+
|
|
|
+df_estimators.loc[(df_estimators.condition == 'awake1'),"condition"] = 'awake'
|
|
|
+
|
|
|
+df_estimators.loc[:,"CONDITION"] = 'post'
|
|
|
+
|
|
|
+df_estimators.loc[(df_estimators.condition == 'awake'),"CONDITION"] = 'awake'
|
|
|
+df_estimators.loc[(df_estimators.condition == 'iso'),"CONDITION"] = 'iso'
|
|
|
+df_estimators.loc[(df_estimators.condition == 'keta'),"CONDITION"] = 'keta'
|
|
|
+df_estimators.loc[(df_estimators.condition == 'mmf'),"CONDITION"] = 'mmf'
|
|
|
+
|
|
|
+print(np.unique(df_estimators["CONDITION"]))
|
|
|
+
|
|
|
+#df_estimators["multihue"] = df_estimators["animal"].astype("string") + df_estimators["CONDITION"]
|
|
|
+
|
|
|
+df_estimators["batch"] = (df_estimators["recording"] > 101).astype("int") + 1
|
|
|
+
|
|
|
+#df_estimators["multihue"] = df_estimators["animal"].astype("string") + df_estimators["CONDITION"]
|
|
|
+
|
|
|
+df_estimators["animal"] = df_estimators["animal"].astype("string")
|
|
|
+
|
|
|
+#df_estimators = df_estimators[df_estimators.animal == 'F1']
|
|
|
+
|
|
|
+print(np.unique(df_estimators["animal"].astype("string")))
|
|
|
+
|
|
|
+df_estimators["multihue"] = "batch" + df_estimators["batch"].astype("string") + df_estimators["CONDITION"]
|
|
|
+
|
|
|
+#print(np.unique(df_estimators["batch"]))
|
|
|
+
|
|
|
+print(np.unique(df_estimators["CONDITION"]))
|
|
|
+
|
|
|
+#print()
|
|
|
+
|
|
|
+
|
|
|
+# ### Plot
|
|
|
+
|
|
|
+# In[9]:
|
|
|
+
|
|
|
+parameters = ['number.neurons','traces.median','traces.skewness','decay','median.stability']
|
|
|
+labels = ['Extracted \n ROIs','Median, \n A.U.','Skewness','Decay time, \n s','1st/2nd \n ratio, %']
|
|
|
+number_subplots = len(parameters)
|
|
|
+
|
|
|
+recordings_ranges = [[0,198]]
|
|
|
+
|
|
|
+#sns.stripplot(x='multihue', y='number.neurons', data=df_estimators, jitter=True)
|
|
|
+
|
|
|
+#sns.scatterplot(x='multihue', y='number.neurons', data=df_estimators)
|
|
|
+
|
|
|
+#plt.show()
|
|
|
+
|
|
|
+#sns.swarmplot(x='multihue', y='number.neurons', data=df_estimators)
|
|
|
+
|
|
|
+
|
|
|
+for rmin,rmax in recordings_ranges:
|
|
|
+
|
|
|
+ f, axes = plt.subplots(number_subplots, 1, figsize=(1, 5)) # sharex=Truerex=True
|
|
|
+ #plt.subplots_adjust(left=None, bottom=0.1, right=None, top=0.9, wspace=None, hspace=0.2)
|
|
|
+ #f.tight_layout()
|
|
|
+ sns.despine(left=True)
|
|
|
+
|
|
|
+ for i, param in enumerate(parameters):
|
|
|
+
|
|
|
+ lw = 0.8
|
|
|
+
|
|
|
+ #else:
|
|
|
+
|
|
|
+ df_nneurons = df_estimators.groupby(['recording','multihue','CONDITION'], as_index=False)['number.neurons'].median()
|
|
|
+
|
|
|
+ if (i == 0):
|
|
|
+ sns.stripplot(x='CONDITION', y='number.neurons', data=df_nneurons[(df_nneurons.recording>=rmin)&(df_nneurons.recording<=rmax)], hue = "CONDITION", palette = custom_palette, order = ['awake', 'iso', 'mmf', 'keta', 'post'], ax=axes[i], marker = '.',edgecolor="black", linewidth = lw*0.5) #
|
|
|
+ else:
|
|
|
+ sns.boxplot(x='CONDITION', y=param, data=df_estimators[(df_estimators.recording>=rmin)&(df_estimators.recording<=rmax)], hue = "CONDITION", palette = custom_palette, dodge=False, order = ['awake', 'iso', 'mmf','keta', 'post' ]
|
|
|
+, showfliers = False,ax=axes[i],linewidth=lw)
|
|
|
+ # param = "animal"
|
|
|
+ # print(np.unique(df_estimators[param]))
|
|
|
+ # axes[i].set_yticks(np.unique(df_estimators[param]))
|
|
|
+
|
|
|
+ # sns.swarmplot(x='recording', y=param, data=df_estimators[(df_estimators.recording>=rmin)&(df_estimators.recording<=rmax)&(df_estimators['neuronID'] == 0)],dodge=False, s=1, edgecolor='black', linewidth=1, ax=axes[i])
|
|
|
+ #ax.set(ylabel="")
|
|
|
+ if i == 0:
|
|
|
+ axes[i].set_ylim([0.0,1200.0])
|
|
|
+ if i > 0:
|
|
|
+ axes[i].set_ylim([0.0,1000.0])
|
|
|
+ if i > 1:
|
|
|
+ axes[i].set_ylim([0.0,10.0])
|
|
|
+ if i > 2:
|
|
|
+ axes[i].set_ylim([0.0,1.0])
|
|
|
+ if i > 3:
|
|
|
+ axes[i].set_ylim([90,110])
|
|
|
+ axes[i].get_xaxis().set_visible(True)
|
|
|
+ else:
|
|
|
+ axes[i].get_xaxis().set_visible(False)
|
|
|
+
|
|
|
+ #if i < number_subplots-1:
|
|
|
+ axes[i].xaxis.label.set_visible(False)
|
|
|
+ #if i==0:
|
|
|
+ # axes[i].set_title("Validation: stability check (recordings #%d-#%d)" % (rmin,rmax), fontsize=9, pad=30) #45
|
|
|
+ axes[i].set_ylabel(labels[i], fontsize=9,labelpad=5) #40
|
|
|
+ #axes[i].set_xlabel("Recording", fontsize=5,labelpad=5) #40
|
|
|
+ #axes[i].axis('off')
|
|
|
+
|
|
|
+ axes[i].xaxis.set_tick_params(labelsize=9) #35
|
|
|
+ axes[i].yaxis.set_tick_params(labelsize=9) #30
|
|
|
+
|
|
|
+ axes[i].get_legend().remove()
|
|
|
+ #axes[i].xaxis.set_major_locator(ticker.MultipleLocator(10))
|
|
|
+ #axes[i].xaxis.set_major_formatter(ticker.ScalarFormatter())
|
|
|
+
|
|
|
+ plt.xticks(rotation=90)
|
|
|
+ #plt.legend(bbox_to_anchor=(1.01, 1), loc=2, borderaxespad=0.,fontsize=25)
|
|
|
+ plt.savefig("Validation_stability_check_figure2_super_compact.png",dpi=400)
|
|
|
+ plt.savefig("Validation_stability_check_figure2_super_compact.svg")
|
|
|
+ #plt.show()
|
|
|
+
|
|
|
+
|
|
|
+# In[13]:
|
|
|
+
|
|
|
+
|
|
|
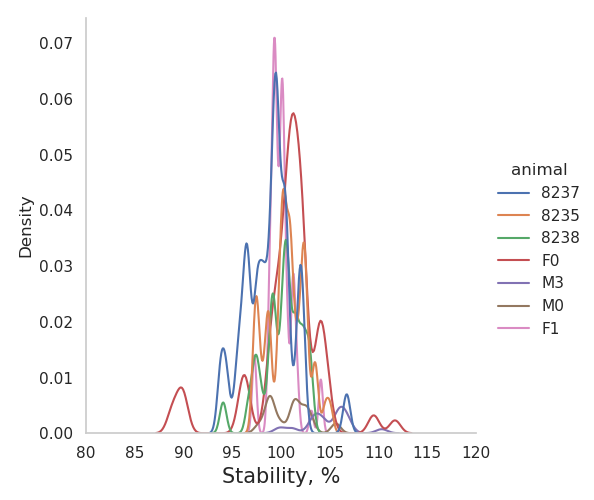
+sns.displot(data=df_estimators, x="median.stability", kind="kde", hue = "animal")
|
|
|
+plt.xlim([80,120])
|
|
|
+plt.xlabel("Stability, %", fontsize = 15)
|
|
|
+plt.title("Validation: summary on stability (recordings #%d-#%d)" % (min(rec),max(rec)), fontsize = 20, pad=20)
|
|
|
+plt.grid(False)
|
|
|
+plt.savefig("Validation_summary_stability_recordings_#%d-#%d)" % (min(rec),max(rec)))
|
|
|
+#plt.show()
|
|
|
+
|
|
|
+
|
|
|
+# In[62]:
|
|
|
+
|
|
|
+
|
|
|
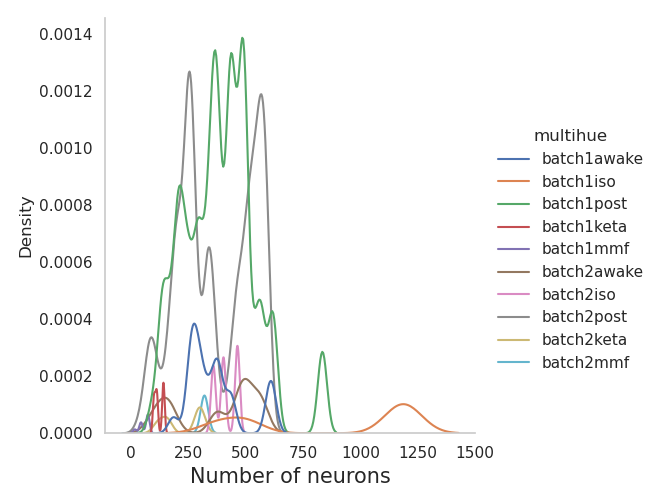
+sns.displot(data=df_estimators, x="number.neurons", kind="kde", hue = "multihue")
|
|
|
+#plt.xlim([80,120])
|
|
|
+plt.xlabel("Number of neurons", fontsize = 15)
|
|
|
+
|
|
|
+plt.grid(False)
|
|
|
+plt.savefig("Number of neurons multihue.png")
|
|
|
+#plt.show()
|
|
|
+
|