|
|
@@ -1,13 +1,13 @@
|
|
|
---
|
|
|
title: Supplementary Materials to Establishing the reliability and validity of measures extracted from long-form recordings
|
|
|
output:
|
|
|
+ pdf_document:
|
|
|
+ toc: yes
|
|
|
+ toc_depth: 3
|
|
|
html_document:
|
|
|
toc: yes
|
|
|
toc_depth: '3'
|
|
|
df_print: paged
|
|
|
- pdf_document:
|
|
|
- toc: yes
|
|
|
- toc_depth: 3
|
|
|
---
|
|
|
|
|
|
```{r setup, include=FALSE, eval=TRUE}
|
|
|
@@ -157,30 +157,30 @@ Third, and perhaps most relevant, we looked for references that evaluated the ps
|
|
|
|
|
|
|
|
|
```{r tab2}
|
|
|
-chi_per_corpus= aggregate(data = mydat_aclew, child_id ~ experiment, function(child_id) length(unique(child_id)))[,2]
|
|
|
+chiXcor= aggregate(data = mydat_aclew, child_id ~ experiment, function(child_id) length(unique(child_id)))[,2]
|
|
|
|
|
|
-rec_per_corpus = aggregate(data = mydat_aclew, session_id ~ experiment, function(session_id) length(unique(session_id)))[,2]
|
|
|
+recXcor = aggregate(data = mydat_aclew, session_id ~ experiment, function(session_id) length(unique(session_id)))[,2]
|
|
|
|
|
|
rec_per_child = setNames(aggregate(data = mydat_aclew, session_id ~ experiment*child_id, function(session_id) length(unique(session_id))), c('experiment', 'Chi', 'No_rec'))
|
|
|
|
|
|
min_rec_per_child = aggregate(data = rec_per_child, No_rec ~ experiment, min)[,2]
|
|
|
max_rec_per_child = aggregate(data = rec_per_child, No_rec ~ experiment, max)[,2]
|
|
|
-rec_r_per_child = paste(min_rec_per_child,max_rec_per_child,sep="-")
|
|
|
+recRXchi = paste(min_rec_per_child,max_rec_per_child,sep="-")
|
|
|
|
|
|
-dur_per_corpus = aggregate(data = mydat_aclew, duration_vtc ~ experiment, function(duration_vtc) round(mean(duration_vtc)/3.6e+6,1))[,2]
|
|
|
+durXcor = aggregate(data = mydat_aclew, duration_vtc ~ experiment, function(duration_vtc) round(mean(duration_vtc)/3.6e+6,1))[,2]
|
|
|
|
|
|
-age_mean_per_corpus = aggregate(data = mydat_aclew, age ~ experiment, function(age) round(mean(age),1))[,2]
|
|
|
+ageXcor = aggregate(data = mydat_aclew, age ~ experiment, function(age) round(mean(age),1))[,2]
|
|
|
|
|
|
age_min_per_corpus = aggregate(data = mydat_aclew, age ~ experiment, function(age) min(age))[,2]
|
|
|
|
|
|
age_max_per_corpus = aggregate(data = mydat_aclew, age ~ experiment, function(age) max(age))[,2]
|
|
|
|
|
|
-age_r_per_corpus = paste(age_min_per_corpus,age_max_per_corpus,sep="-")
|
|
|
+ageRXcor = paste(age_min_per_corpus,age_max_per_corpus,sep="-")
|
|
|
|
|
|
corpus=c("bergelson", "cougar", "fausey-trio", "lucid","lyon", "quechua", "warlaumont", "winnipeg")
|
|
|
location=c("Northeast US", "Northwest US", "Western US", "Northwest England", "Central France", "Highlands Bolivia", "Western US", "Western Canada")
|
|
|
|
|
|
-corpus_description=cbind(corpus,location,chi_per_corpus, rec_r_per_child, rec_per_corpus, dur_per_corpus, age_mean_per_corpus,age_r_per_corpus)
|
|
|
+corpus_description=cbind(corpus,location,chiXcor, recRXchi, recXcor, durXcor, ageXcor,ageRXcor)
|
|
|
|
|
|
write.table(corpus_description, "../output/corpus_description.csv", sep='\t')
|
|
|
|
|
|
@@ -195,7 +195,7 @@ nrecs=length(levels(mydat_aclew$session_id))
|
|
|
|
|
|
## SM D: Code to reproduce Fig. 2
|
|
|
|
|
|
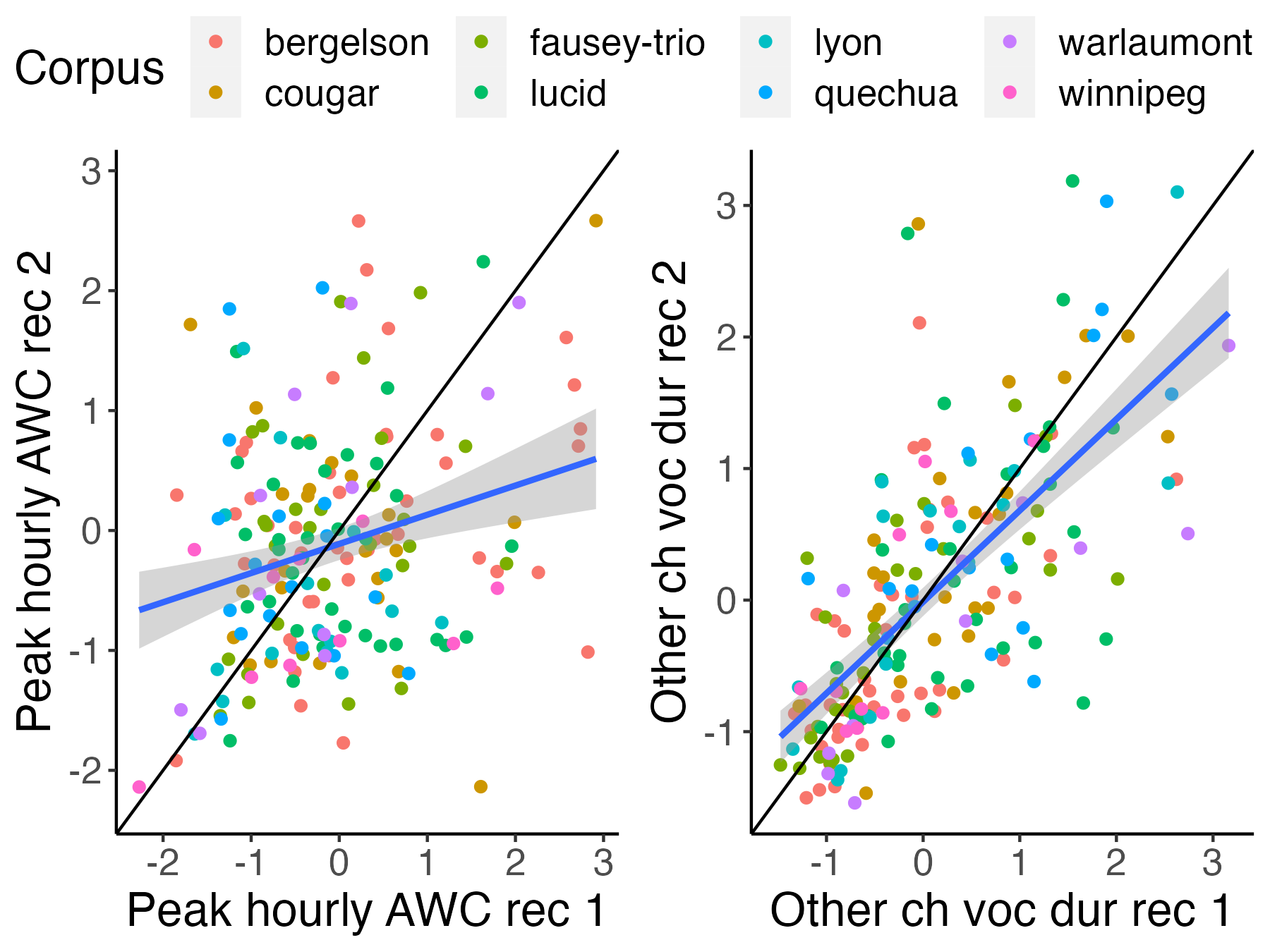
-```{r icc-examples-fig2, fig.width=4, fig.height=3,fig.cap="Figure 2 (reproduced). Scatterplots for two selected variables. The left one has relatively low ICCs; the right one has relatively higher ICCs."}
|
|
|
+```{r icc-examples-fig2, fig.width=6, fig.height=4.5,fig.cap="Figure 2 (reproduced). Scatterplots for two selected variables. The left one has relatively low ICCs; the right one has relatively higher ICCs."}
|
|
|
# figure of bad ICC: lena used to be: avg_voc_dur_chi, now is: peak_wc_adu_ph; good ICC: lena used to be: voc_och_ph, now is: voc_dur_och_ph
|
|
|
|
|
|
# remove missing data points altogether
|
|
|
@@ -258,11 +258,12 @@ panel.background = element_blank(), axis.line = element_line(colour = "black"))
|
|
|
geom_abline(intercept = 0, slope = 1)
|
|
|
|
|
|
|
|
|
-ggarrange(bad, good,
|
|
|
+fig2 = ggarrange(bad, good,
|
|
|
ncol = 2, nrow = 1, common.legend = TRUE, vjust = 1.5, hjust=0,
|
|
|
font.label = list(size = 20)) + labs(color= "Corpus") + theme(text = element_text(size = 20))
|
|
|
+fig2
|
|
|
|
|
|
-
|
|
|
+ggsave("fig2.png", plot = fig2, width = 6, height = 4.5, units = "in")
|
|
|
```
|
|
|
|
|
|
## SM E: Code to reproduce text at the beginning of the "Setting the stage" section
|
|
|
@@ -318,7 +319,7 @@ cor_t=t.test(rval_tab$m ~ rval_tab$data_set)
|
|
|
|
|
|
```
|
|
|
|
|
|
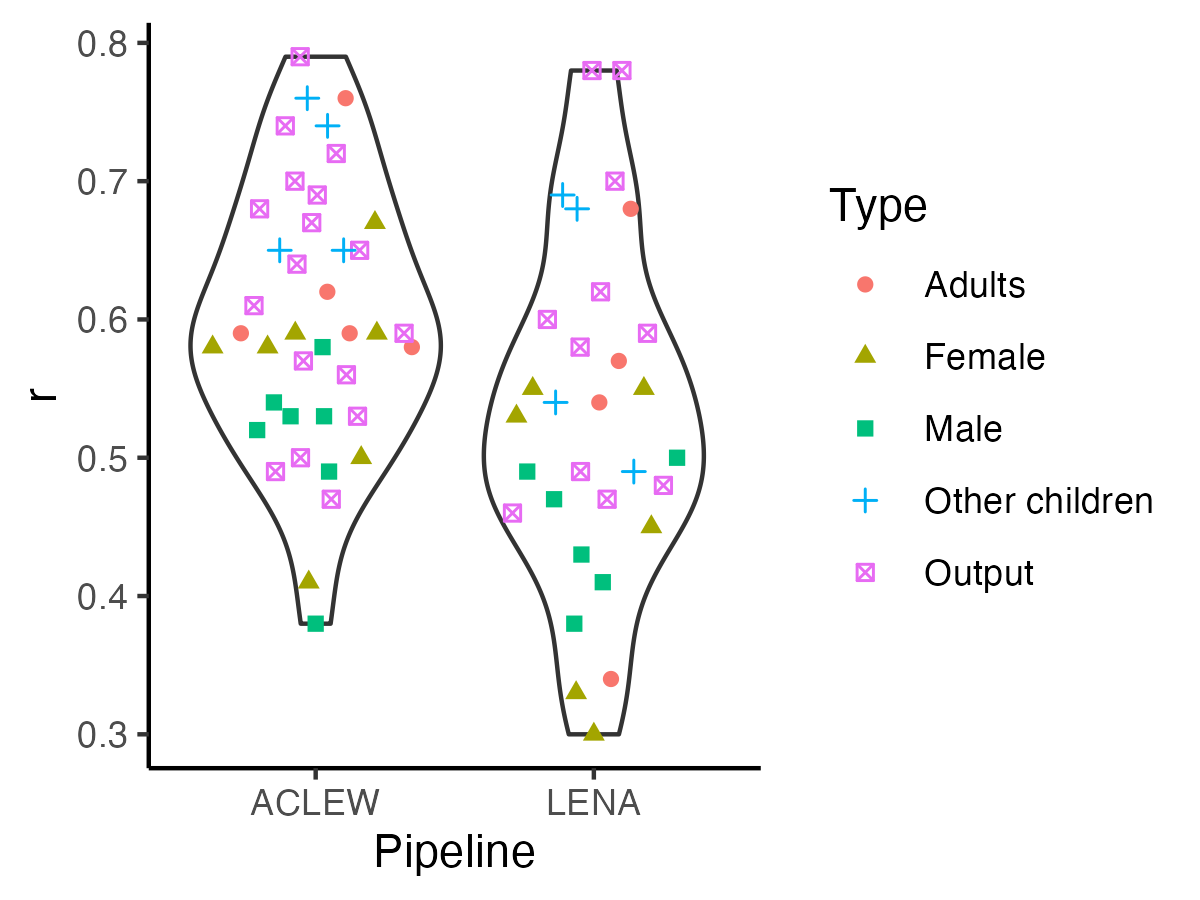
-> To see whether correlations in this analysis differed by talker types and pipelines, we fit a linear model with the formula $lm(cor ~ type * pipeline)$, where type indicates whether the measure pertained to the key child, (female/male) adults, other children; and pipeline LENA or ACLEW. The model was overall significant (F(`round(reg_sum_cor$fstatistic["dendf"],2)`) = `round(reg_sum_cor$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum_cor$adj.r.squared*100)`%, suggesting this model did not explain a great deal of variance in correlation coefficients. A Type 3 ANOVA on this model revealed a significant effect of pipeline (F = `r round(reg_anova_cor["data_set","F value"],2)`, p = `r round(reg_anova_cor["data_set","Pr(>F)"],2)`), due to higher correlations for ACLEW (`r r_msds["aclew","x"]`) than for LENA metrics (m = `r r_msds["lena","x"]`).
|
|
|
+> To see whether correlations in this analysis differed by talker types and pipelines, we fit a linear model with the formula $lm(cor ~ type * pipeline)$, where type indicates whether the measure pertained to the key child, (female/male) adults, other children; and pipeline LENA or ACLEW. The model was overall significant (F(`r round(reg_sum_cor$fstatistic["dendf"],2)`) = `r round(reg_sum_cor$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum_cor$adj.r.squared*100)`%, suggesting this model did not explain a great deal of variance in correlation coefficients. A Type 3 ANOVA on this model revealed a significant effect of pipeline (F = `r round(reg_anova_cor["data_set","F value"],2)`, p = `r round(reg_anova_cor["data_set","Pr(>F)"],2)`), due to higher correlations for ACLEW (`r r_msds["aclew","x"]`) than for LENA metrics (m = `r r_msds["lena","x"]`).
|
|
|
|
|
|
See table below for results of the Type 3 ANOVA.
|
|
|
|
|
|
@@ -333,11 +334,16 @@ kable(round(reg_anova_cor,2),caption="Type 3 ANOVA on model attempting to explai
|
|
|
```{r r-fig4, echo=F,fig.width=4, fig.height=3,fig.cap="Figure 4 (reproduced). Violin plot reflecting the distribution of correlations."}
|
|
|
|
|
|
|
|
|
-ggplot(rval_tab, aes(y = m, x = toupper(data_set))) +
|
|
|
+fig4 <- ggplot(rval_tab, aes(y = m, x = toupper(data_set))) +
|
|
|
geom_violin(alpha = 0.5) +
|
|
|
geom_quasirandom(aes(colour = Type,shape = Type)) +
|
|
|
- theme() +labs( y = "r",x="Pipeline")
|
|
|
+ theme() +labs( y = "r",x="Pipeline") +
|
|
|
+ theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
|
|
|
+panel.background = element_blank(), legend.key=element_blank(), axis.line = element_line(colour = "black"))
|
|
|
|
|
|
+fig4
|
|
|
+
|
|
|
+ggsave("fig4.png", plot = fig4, width = 4, height = 3, units = "in")
|
|
|
|
|
|
```
|
|
|
|
|
|
@@ -446,13 +452,17 @@ panel.background = element_blank(), legend.key=element_blank(), axis.line = elem
|
|
|
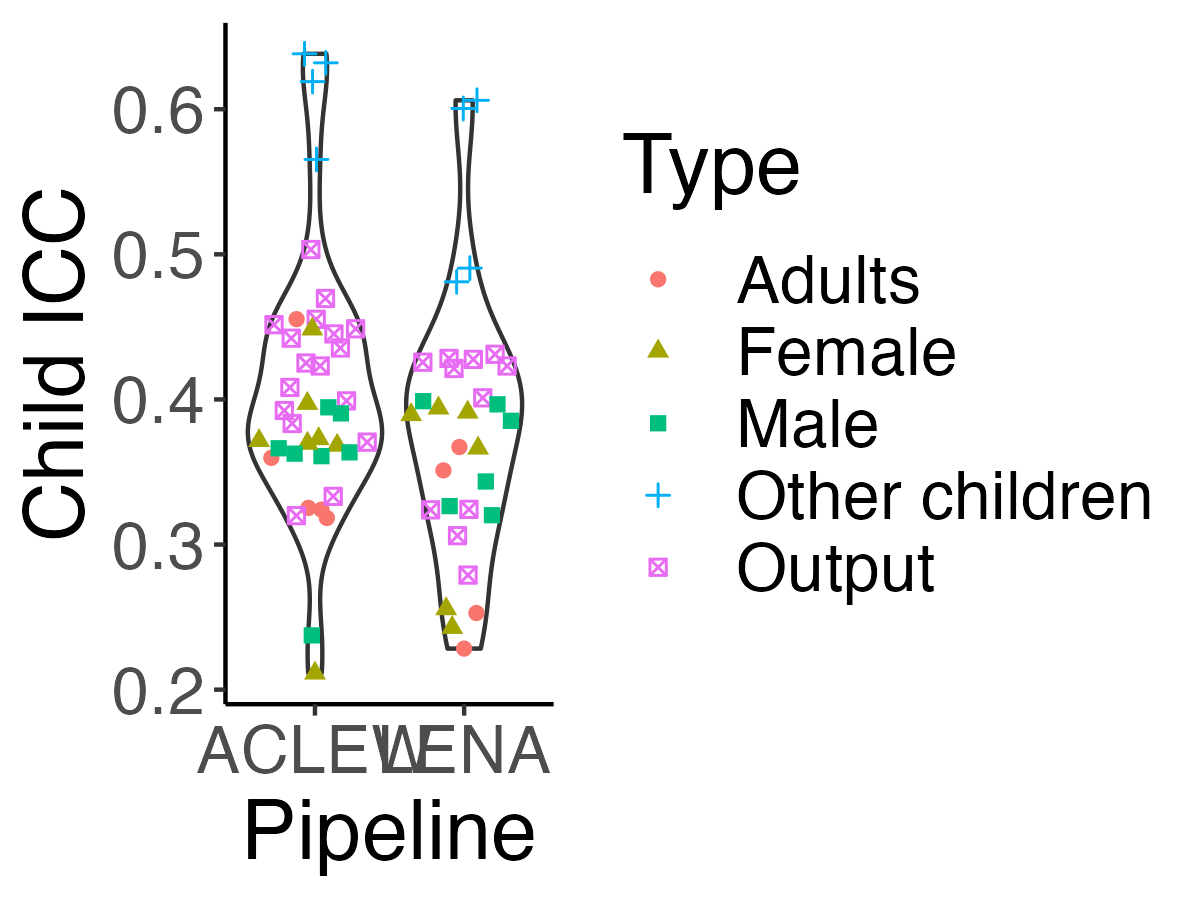
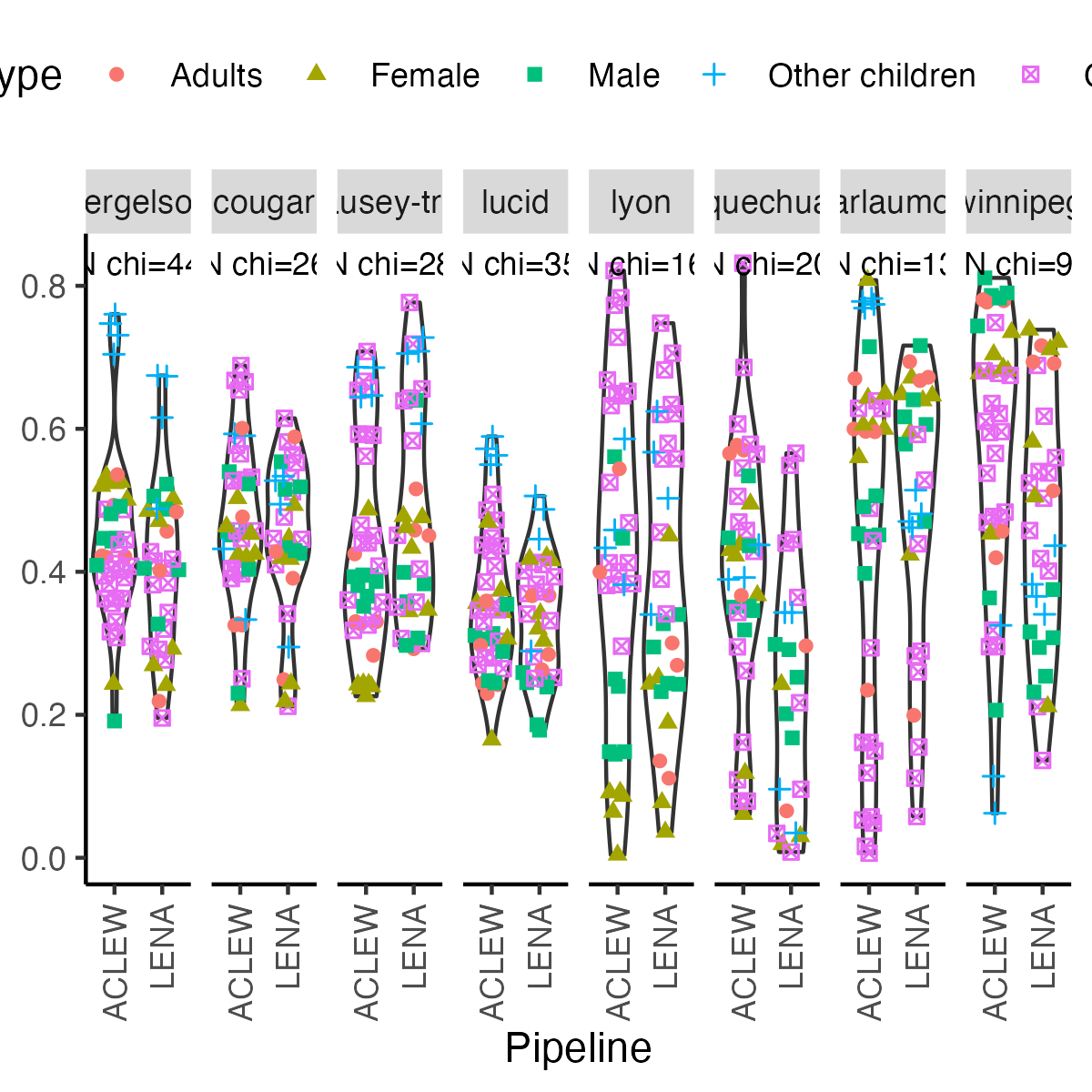
```{r icc-allexp-fig5, echo=F,fig.width=4, fig.height=3,fig.cap="Figure 5 (reproduced). Violin plot reflecting the distribution of Child ICC."}
|
|
|
|
|
|
|
|
|
-ggplot(df.icc.mixed, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
+fig5 <- ggplot(df.icc.mixed, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
geom_violin(alpha = 0.5) +
|
|
|
geom_quasirandom(aes(colour = Type,shape = Type)) +
|
|
|
labs( y = "Child ICC",x="Pipeline") + theme(text = element_text(size = 20)) +
|
|
|
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
|
|
|
panel.background = element_blank(), legend.key=element_blank(), axis.line = element_line(colour = "black"))
|
|
|
|
|
|
+fig5
|
|
|
+
|
|
|
+ggsave("fig5.png", plot = fig5, width = 4, height = 3, units = "in")
|
|
|
+
|
|
|
```
|
|
|
|
|
|
|
|
|
@@ -478,7 +488,7 @@ rownames(msds_p)<-msds_p$Group.1
|
|
|
```
|
|
|
|
|
|
|
|
|
-> Next, we explored how similar Child ICCs were across different talker types and pipelines. We fit a linear model with the formula $lm(icc\_child\_id ~ type * pipeline)$, where type indicates whether the measure pertained to the key child, (female/male) adults, other children; and pipeline LENA or ACLEW. The model was overall significant (F(`round(reg_sum$fstatistic["dendf"],2)`) = `round(reg_sum$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum$adj.r.squared*100)`%, suggesting much of the variance across Child ICCs was explained by these factors. A Type 3 ANOVA on this model revealed type was a signficant predictor (F(`r reg_anova["Type","Df"]`) = `r round(reg_anova["Type","F value"],1)`, p<.001), as was pipeline (F(`r reg_anova["data_set","Df"]`) = `r round(reg_anova["data_set","F value"],1)`, p = `r round(reg_anova["data_set","Pr(>F)"],3)`); the interaction between type and pipeline was not significant. The main effect of type emerged because output metrics tended to have higher Child ICC (`r msds["Output","x"]`) than those associated to adults in general (`r msds["Adults","x"]`), females (`r msds["Female","x"]`), and males (`r msds["Male","x"]`); whereas those associated with other children had even higher Child ICCs (`r msds["Other children","x"]`). The main effect of pipeline arose because of slightly higher Child ICCs for the ACLEW metrics (`r msds_p["aclew","x"]`) than for LENA metrics (`r msds_p["lena","x"]`).
|
|
|
+> Next, we explored how similar Child ICCs were across different talker types and pipelines. We fit a linear model with the formula $lm(icc\_child\_id ~ type * pipeline)$, where type indicates whether the measure pertained to the key child, (female/male) adults, other children; and pipeline LENA or ACLEW. The model was overall significant (F(`r round(reg_sum$fstatistic["dendf"],2)`) = `r round(reg_sum$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum$adj.r.squared*100)`%, suggesting much of the variance across Child ICCs was explained by these factors. A Type 3 ANOVA on this model revealed type was a signficant predictor (F(`r reg_anova["Type","Df"]`) = `r round(reg_anova["Type","F value"],1)`, p<.001), as was pipeline (F(`r reg_anova["data_set","Df"]`) = `r round(reg_anova["data_set","F value"],1)`, p = `r round(reg_anova["data_set","Pr(>F)"],3)`); the interaction between type and pipeline was not significant. The main effect of type emerged because output metrics tended to have higher Child ICC (`r msds["Output","x"]`) than those associated to adults in general (`r msds["Adults","x"]`), females (`r msds["Female","x"]`), and males (`r msds["Male","x"]`); whereas those associated with other children had even higher Child ICCs (`r msds["Other children","x"]`). The main effect of pipeline arose because of slightly higher Child ICCs for the ACLEW metrics (`r msds_p["aclew","x"]`) than for LENA metrics (`r msds_p["lena","x"]`).
|
|
|
|
|
|
|
|
|
## SM O: Code to reproduce Table 4
|
|
|
@@ -528,7 +538,7 @@ f_labels<-data.frame(age_bin=levels(df.icc.age$age_bin),facet_labels_chi=facet_l
|
|
|
|
|
|
f_labels$age_bin<-factor(f_labels$age_bin,levels=age_levels)
|
|
|
|
|
|
-ggplot(df.icc.age, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
+fig6 <- ggplot(df.icc.age, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
geom_violin(alpha = 0.5) +
|
|
|
geom_quasirandom(aes(colour = Type,shape = Type)) +
|
|
|
theme(legend.position="none") +labs( y = "r",x="Pipeline") + facet_wrap(~age_bin, ncol = 3) +
|
|
|
@@ -537,6 +547,9 @@ ggplot(df.icc.age, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
|
|
|
panel.background = element_blank(), legend.key=element_blank(), axis.line = element_line(colour = "black"))
|
|
|
|
|
|
+fig6
|
|
|
+
|
|
|
+ggsave("fig6.png", plot = fig6, width = 6, height = 10, units = "in")
|
|
|
|
|
|
|
|
|
```
|
|
|
@@ -556,7 +569,7 @@ reg_anova_age_icc=Anova(age_icc)
|
|
|
|
|
|
```
|
|
|
|
|
|
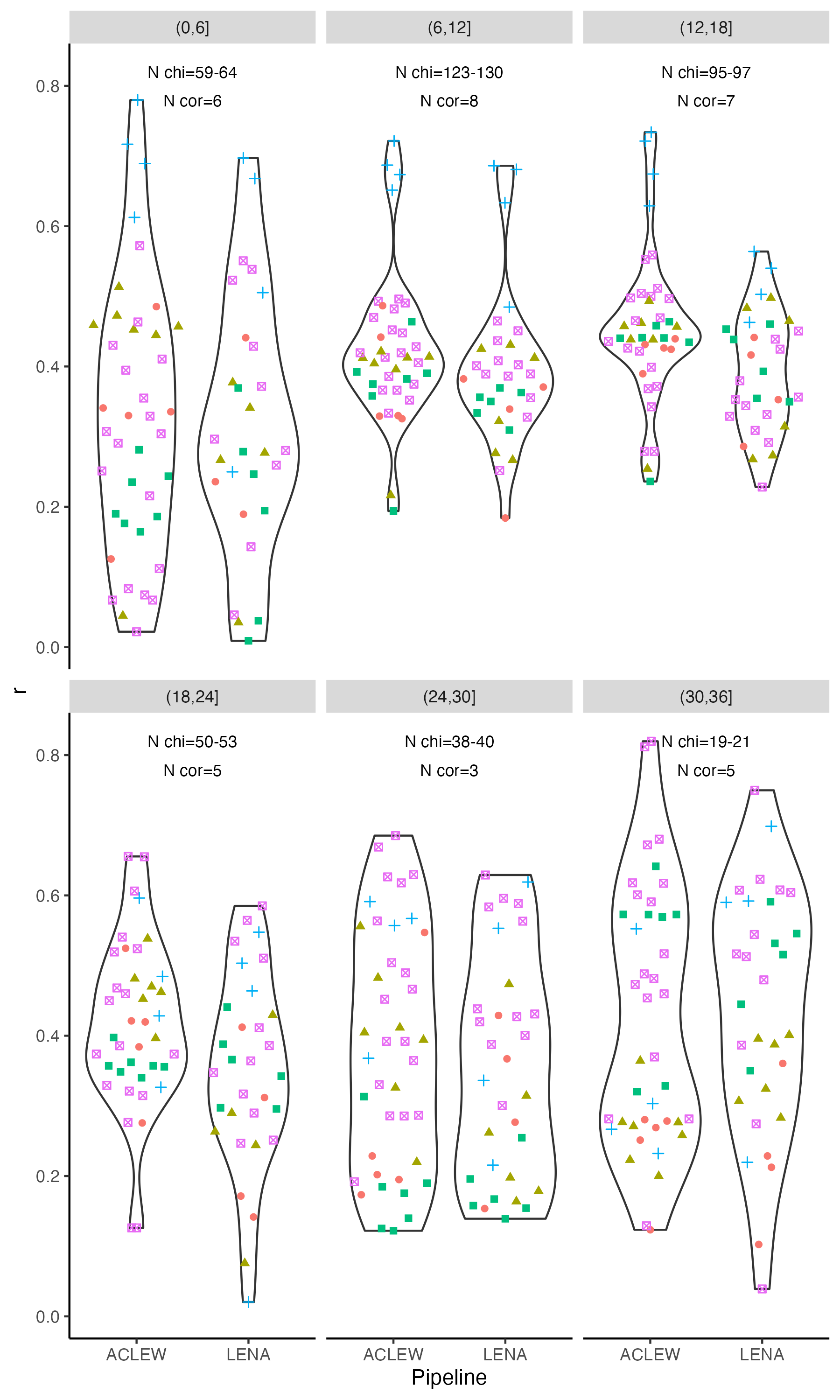
-> To interrogate these results statistically, and assess whether Child ICCs tended to be higher or lower in certain age bins, we fit a linear model with the formula $lm(Child_ICC ~ type * pipeline * age_bin)$. The model was overall significant (F(`round(reg_sum_age_icc$fstatistic["dendf"],2)`) = `round(reg_sum_age_icc$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum_age_icc$adj.r.squared*100)`%, suggesting this model explained about a third of the variance in Child ICC. A Type 3 ANOVA on this model revealed type was a signficant predictor (F(`r reg_anova["Type","Df"]`) = `r round(reg_anova["Type","F value"],1)`, p<.001), whereas as was pipeline (F(`r reg_anova["data_set","Df"]`) = `r round(reg_anova["data_set","F value"],1)`, p = `r round(reg_anova["data_set","Pr(>F)"],3)`); the interaction between type and pipeline was not significant.
|
|
|
+> To interrogate these results statistically, and assess whether Child ICCs tended to be higher or lower in certain age bins, we fit a linear model with the formula $lm(Child_ICC ~ type * pipeline * age_bin)$. The model was overall significant (F(`r round(reg_sum_age_icc$fstatistic["dendf"],2)`) = `r round(reg_sum_age_icc$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum_age_icc$adj.r.squared*100)`%, suggesting this model explained about a third of the variance in Child ICC. A Type 3 ANOVA on this model revealed type was a signficant predictor (F(`r reg_anova["Type","Df"]`) = `r round(reg_anova["Type","F value"],1)`, p<.001), whereas as was pipeline (F(`r reg_anova["data_set","Df"]`) = `r round(reg_anova["data_set","F value"],1)`, p = `r round(reg_anova["data_set","Pr(>F)"],3)`); the interaction between type and pipeline was not significant.
|
|
|
|
|
|
See table below for results of the Type 3 ANOVA.
|
|
|
|
|
|
@@ -591,12 +604,16 @@ r_X_age$ageA=factor(r_X_age$ageA,levels=age_levels)
|
|
|
|
|
|
#summary(r_X_age$cor) #mean correlation across corpora is zero!
|
|
|
|
|
|
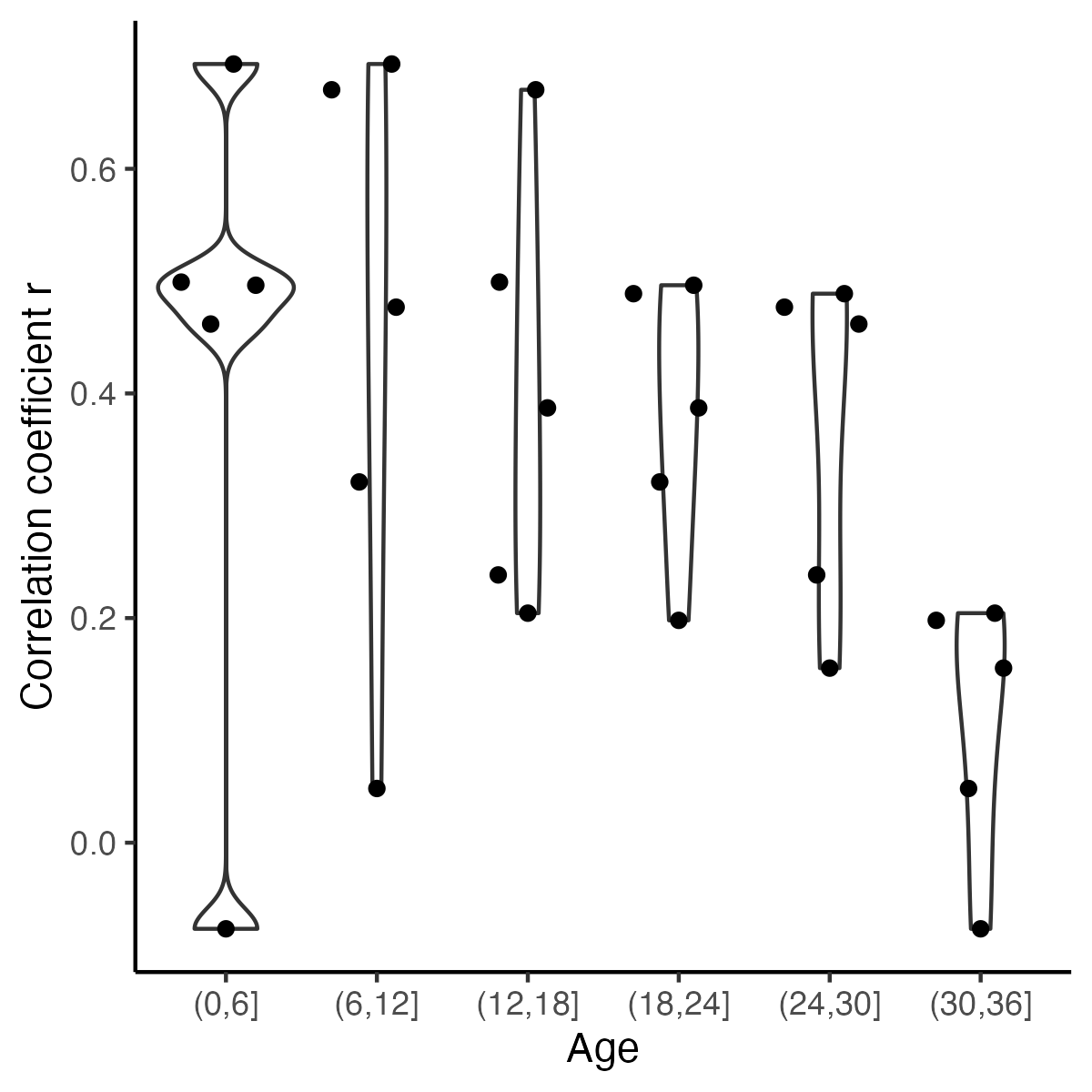
-ggplot(r_X_age, aes(y = cor, x = ageA)) +
|
|
|
+fig7 <- ggplot(r_X_age, aes(y = cor, x = ageA)) +
|
|
|
geom_violin(alpha = 0.5) +
|
|
|
geom_quasirandom() +
|
|
|
theme() +labs( y = "Correlation coefficient r",x="Age") +
|
|
|
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
|
|
|
panel.background = element_blank(), legend.key=element_blank(), axis.line = element_line(colour = "black"))
|
|
|
+
|
|
|
+fig7
|
|
|
+
|
|
|
+ggsave("fig7.png", plot = fig7, width = 4, height = 4, units = "in")
|
|
|
```
|
|
|
|
|
|
|
|
|
@@ -610,16 +627,16 @@ panel.background = element_blank(), legend.key=element_blank(), axis.line = elem
|
|
|
## SM U: Code to reproduce Figure 8
|
|
|
|
|
|
|
|
|
-```{r icc-bycor-fig8, echo=F,fig.width=4, fig.height=10,fig.cap="Figure 8 (reproduced). Child ICC by metric type and pipeline, when considering each corpus separately."}
|
|
|
+```{r icc-bycor-fig8, echo=F,fig.width=4, fig.height=4,fig.cap="Figure 8 (reproduced). Child ICC by metric type and pipeline, when considering each corpus separately."}
|
|
|
|
|
|
-facet_labels_chi = paste0("N chi=",chi_per_corpus)
|
|
|
+facet_labels_chi = paste0("N chi=",chiXcor)
|
|
|
|
|
|
#and then we structure it so that it goes on the plot
|
|
|
f_labels<-data.frame(levels(factor(df.icc.corpus$corpus)),facet_labels_chi=facet_labels_chi)
|
|
|
|
|
|
colnames(f_labels)<-c("corpus","nchi")
|
|
|
|
|
|
-ggplot(df.icc.corpus, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
+fig8 <- ggplot(df.icc.corpus, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
geom_violin(alpha = 0.5) +
|
|
|
geom_quasirandom(aes(colour = Type,shape = Type)) +
|
|
|
theme(legend.position = "top", axis.title.y=element_blank() ,axis.text.x = element_text(angle = 90, vjust = 0.5, hjust=1)) +labs( y = "Child ICC",x="Pipeline") +
|
|
|
@@ -628,6 +645,9 @@ ggplot(df.icc.corpus, aes(y = icc_child_id, x = toupper(data_set))) +
|
|
|
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
|
|
|
panel.background = element_blank(), legend.key=element_blank(), axis.line = element_line(colour = "black"))
|
|
|
|
|
|
+fig8
|
|
|
+
|
|
|
+ggsave("fig8.png", plot = fig8, width = 4, height = 4, units = "in")
|
|
|
|
|
|
```
|
|
|
|
|
|
@@ -646,7 +666,7 @@ reg_anova_cor_icc=Anova(cor_icc)
|
|
|
|
|
|
```
|
|
|
|
|
|
-> The fact that we cannot infer reliability from one corpus based on another one was confirmed statistically: We checked whether Child ICC differed by talker types and pipelines across corpora by fitting a linear model with the formula $lm(Child_ICC ~ type * pipeline * corpus)$, where type indicates whether the measure pertained to the key child, (female/male) adults, other children; pipeline LENA or ACLEW; and corpus the corpus ID. The model was overall significant (F(`round(reg_sum_cor_icc$fstatistic["dendf"],2)`) = `round(reg_sum_cor_icc$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum_cor_icc$adj.r.squared*100)`%, suggesting this model explained nearly half of the variance in Child ICC. A Type 3 ANOVA on this model revealed several significant effects and interactions, including a three-way interaction of type, pipeline, and corpus (F(`r reg_anova_cor_icc["Type:data_set:corpus","Df"]`) = `r round(reg_anova_cor_icc["Type:data_set:corpus","F value"],1)`, p<.001); a two-way interaction of type and corpus (F(`r reg_anova_cor_icc["data_set:corpus","Df"]`) = `r round(reg_anova_cor_icc["data_set:corpus","F value"],1)`, p<.001); and a main effect of corpus (F(`r reg_anova_cor_icc["corpus","Df"]`) = `r round(reg_anova_cor_icc["corpus","F value"],1)`, p<.001).
|
|
|
+> The fact that we cannot infer reliability from one corpus based on another one was confirmed statistically: We checked whether Child ICC differed by talker types and pipelines across corpora by fitting a linear model with the formula $lm(Child_ICC ~ type * pipeline * corpus)$, where type indicates whether the measure pertained to the key child, (female/male) adults, other children; pipeline LENA or ACLEW; and corpus the corpus ID. The model was overall significant (F(`r round(reg_sum_cor_icc$fstatistic["dendf"],2)`) = `r round(reg_sum_cor_icc$fstatistic["value"],2)`, p < .001). We found an adjusted R-squared of `r round(reg_sum_cor_icc$adj.r.squared*100)`%, suggesting this model explained nearly half of the variance in Child ICC. A Type 3 ANOVA on this model revealed several significant effects and interactions, including a three-way interaction of type, pipeline, and corpus (F(`r reg_anova_cor_icc["Type:data_set:corpus","Df"]`) = `r round(reg_anova_cor_icc["Type:data_set:corpus","F value"],1)`, p<.001); a two-way interaction of type and corpus (F(`r reg_anova_cor_icc["data_set:corpus","Df"]`) = `r round(reg_anova_cor_icc["data_set:corpus","F value"],1)`, p<.001); and a main effect of corpus (F(`r reg_anova_cor_icc["corpus","Df"]`) = `r round(reg_anova_cor_icc["corpus","F value"],1)`, p<.001).
|
|
|
|
|
|
See Table below for results of the Type 3 ANOVA.
|
|
|
|
|
|
@@ -658,7 +678,7 @@ kable(round(reg_anova_cor_icc,2),caption="Type 3 ANOVA on model attempting to ex
|
|
|
|
|
|
## SM W: Code to reproduce Figure 9
|
|
|
|
|
|
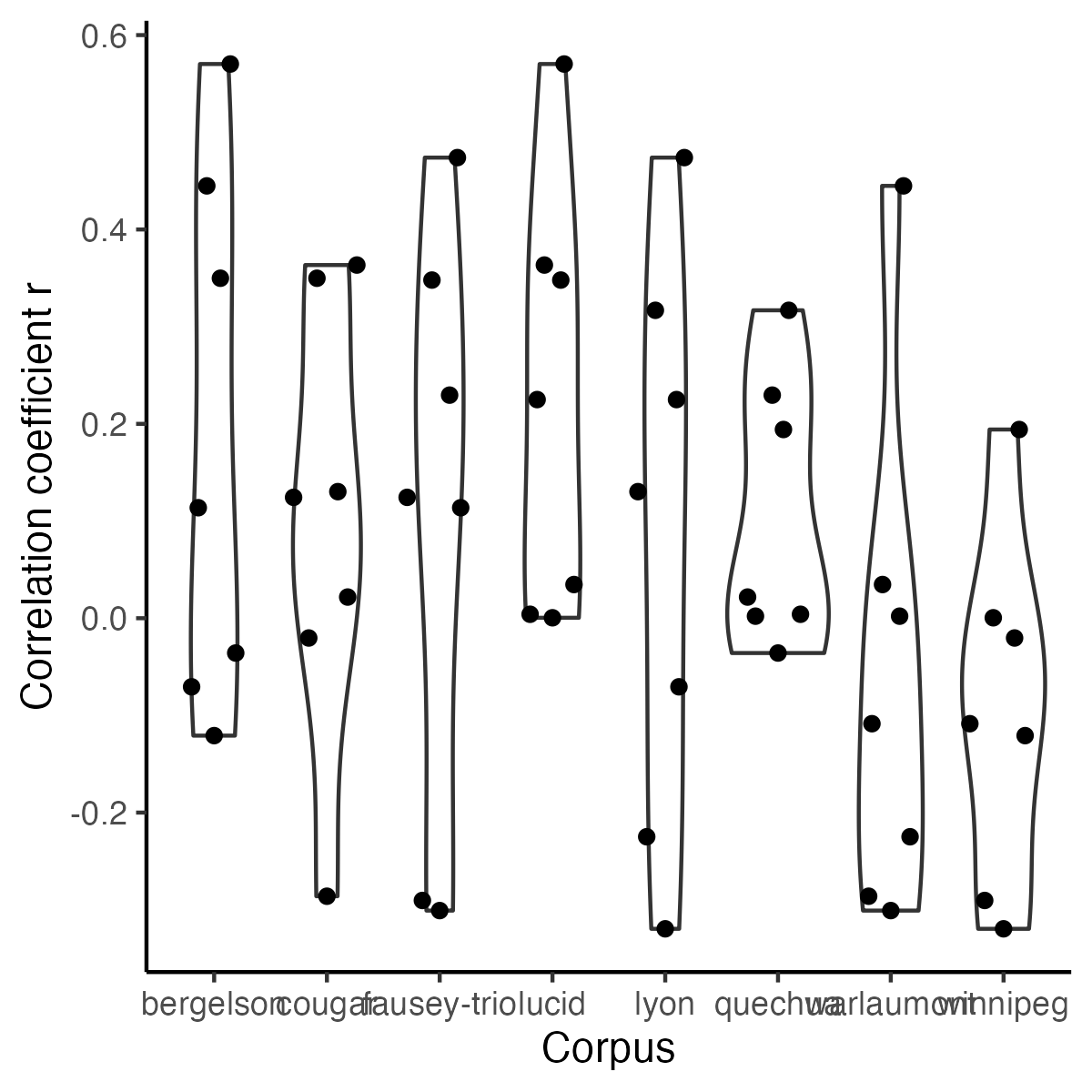
-```{r icc-bycor-fig9, echo=F,fig.width=4, fig.height=10,fig.cap="Figure 9 (reproduced). Correlations in Child ICC across corpora."}
|
|
|
+```{r icc-bycor-fig9, echo=F,fig.width=4, fig.height=4,fig.cap="Figure 9 (reproduced). Correlations in Child ICC across corpora."}
|
|
|
|
|
|
|
|
|
|
|
|
@@ -678,12 +698,16 @@ r_X_corpus$cor=as.numeric(as.character(r_X_corpus$cor))
|
|
|
|
|
|
#summary(r_X_corpus$cor) #mean correlation across corpora is zero!
|
|
|
|
|
|
-ggplot(r_X_corpus, aes(y = cor, x = corpusA)) +
|
|
|
+fig9 <- ggplot(r_X_corpus, aes(y = cor, x = corpusA)) +
|
|
|
geom_violin(alpha = 0.5) +
|
|
|
geom_quasirandom() +
|
|
|
theme() +labs( y = "Correlation coefficient r",x="Corpus") +
|
|
|
theme(panel.grid.major = element_blank(), panel.grid.minor = element_blank(),
|
|
|
panel.background = element_blank(), legend.key=element_blank(), axis.line = element_line(colour = "black"))
|
|
|
+
|
|
|
+fig9
|
|
|
+
|
|
|
+ggsave("fig9.png", plot = fig9, width = 4, height = 4, units = "in")
|
|
|
```
|
|
|
|
|
|
## SM X: Code to reproduce text in the Discussion section
|
|
|
@@ -703,14 +727,14 @@ northam[grep("Bolivia",location)]<-F
|
|
|
northam[grep("France",location)]<-F
|
|
|
northam[grep("England",location)]<-F
|
|
|
|
|
|
-bias_tab<-data.frame(cbind(chi_per_corpus, rec_per_corpus))
|
|
|
-bias_tab$chi_per_corpus<-bias_tab$chi_per_corpus/sum(bias_tab$chi_per_corpus)
|
|
|
-bias_tab$rec_per_corpus<-bias_tab$rec_per_corpus/sum(bias_tab$rec_per_corpus)
|
|
|
+bias_tab<-data.frame(cbind(chiXcor, recXcor))
|
|
|
+bias_tab$chiXcor<-bias_tab$chiXcor/sum(bias_tab$chiXcor)
|
|
|
+bias_tab$recXcor<-bias_tab$recXcor/sum(bias_tab$recXcor)
|
|
|
|
|
|
|
|
|
```
|
|
|
|
|
|
-> Our data draws mainly from urban (`r round(sum(bias_tab$rec_per_corpus[urban])*100)`% of recordings, `r round(sum(bias_tab$chi_per_corpus[urban])*100)`% of the children, `r round(sum(urban)/length(urban)*100)`% of the corpora), English-speaking settings (`r round(sum(bias_tab$rec_per_corpus[english])*100)`% of recordings, `r round(sum(bias_tab$chi_per_corpus[english])*100)`% of the children, `r round(sum(english)/length(english)*100)`% of the corpora), and almost exclusively from North America (`r round(sum(bias_tab$rec_per_corpus[northam])*100)`% of recordings, `r round(sum(bias_tab$chi_per_corpus[northam])*100)`% of the children, `r round(sum(northam)/length(northam)*100)`% of the corpora).
|
|
|
+> Our data draws mainly from urban (`r round(sum(bias_tab$recXcor[urban])*100)`% of recordings, `r round(sum(bias_tab$chiXcor[urban])*100)`% of the children, `r round(sum(urban)/length(urban)*100)`% of the corpora), English-speaking settings (`r round(sum(bias_tab$recXcor[english])*100)`% of recordings, `r round(sum(bias_tab$chiXcor[english])*100)`% of the children, `r round(sum(english)/length(english)*100)`% of the corpora), and almost exclusively from North America (`r round(sum(bias_tab$recXcor[northam])*100)`% of recordings, `r round(sum(bias_tab$chiXcor[northam])*100)`% of the children, `r round(sum(northam)/length(northam)*100)`% of the corpora).
|
|
|
|
|
|
## SM Y: Variability as a function of hardware
|
|
|
|