Se han modificado 42 ficheros con 3830 adiciones y 19 borrados
La diferencia del archivo ha sido suprimido porque es demasiado grande
+ 1843
- 0
code/.ipynb_checkpoints/Big_data_mouse_practical_2020_answer-checkpoint.ipynb
La diferencia del archivo ha sido suprimido porque es demasiado grande
+ 1748
- 0
code/Big_data_mouse_practical_2020_answer.ipynb
+ 103
- 0
code/mouse_connectivity/manifest.json
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 19
- 19
code/overlayPlot.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 109
- 0
code/overlayPlot3D.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
BIN
input/12_wks_coronal_100141780_100um_projection_density.nii.gz
BIN
input/12_wks_coronal_100141780_50um_projection_density.nii.gz
BIN
input/C57BL6_mouse_fiber.nii.gz
BIN
input/SSp_bfd5_Primary_somatosensory_area_barrel_field_layer_5.nii.gz
BIN
input/average_template_100_from_website.nii.gz
BIN
input/average_template_50_from_website.nii.gz
BIN
input/del/1-s2.0-S1053811923001453-gr1.jpg
BIN
input/del/12_wks_coronal_100141780_100um_projection_density.nii.gz
input/12_wks_coronal_100141780_100um_projection_density.nrrd → input/del/12_wks_coronal_100141780_100um_projection_density.nrrd
BIN
input/del/12_wks_coronal_100141780_50um_projection_density.nrrd
BIN
input/del/ABtempate_adj_reg.nii.gz
BIN
input/del/ABtempate_reg.nii.gz
BIN
input/del/ABtemplate100_adjusted.nii.gz
input/ABtemplate_Website.nii.gz → input/del/ABtemplate_Website.nii.gz
BIN
input/del/ABtemplate_Website_registered.nii.gz
BIN
input/del/ABtemplate_adjusted.nii.gz
BIN
input/del/ABtemplate_test.nii.gz
+ 4
- 0
input/del/AffineTraicing.txt
|
||
|
||
|
||
|
||
|
||
BIN
input/del/C57BL6_mouse.fib.image.jpg

+ 4
- 0
input/del/affine4tracing
|
||
|
||
|
||
|
||
|
||
BIN
input/del/average_template_100.nii.gz
BIN
input/del/average_template_100.nrrd
BIN
input/del/average_template_100_adjusted.nii.gz
BIN
input/del/average_template_50.nrrd
BIN
input/del/average_template_50_Niklas.nii.gz
BIN
input/fiber_registered.nii.gz
BIN
output/Nifti_Trakte/CC_Mop_dilated_denstitymask.nii.gz.nii
BIN
output/Nifti_Trakte_registered/CC_Mop_desnitymask_50um.nii.gz
BIN
output/figures/Figure 2024-01-26 114119.png
BIN
output/figures/Figure 2024-01-26 114922.png
BIN
output/figures/Figure 2024-01-26 115238.png

BIN
output/figures/Figure 2024-01-26 115310.png

BIN
output/figures/Figure 2024-01-26 115426.png
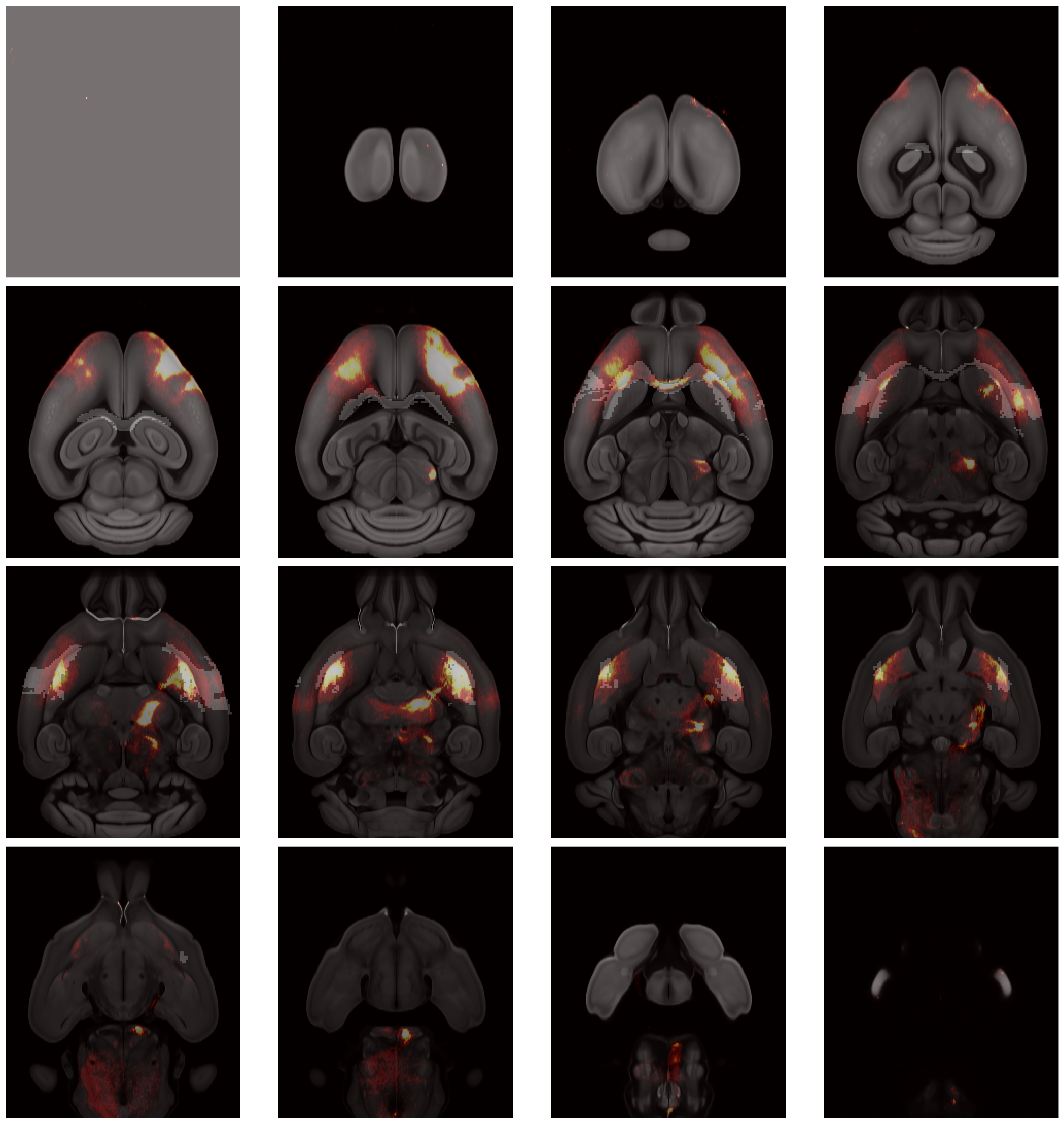
BIN
output/figures/Figure 2024-01-26 132643.png
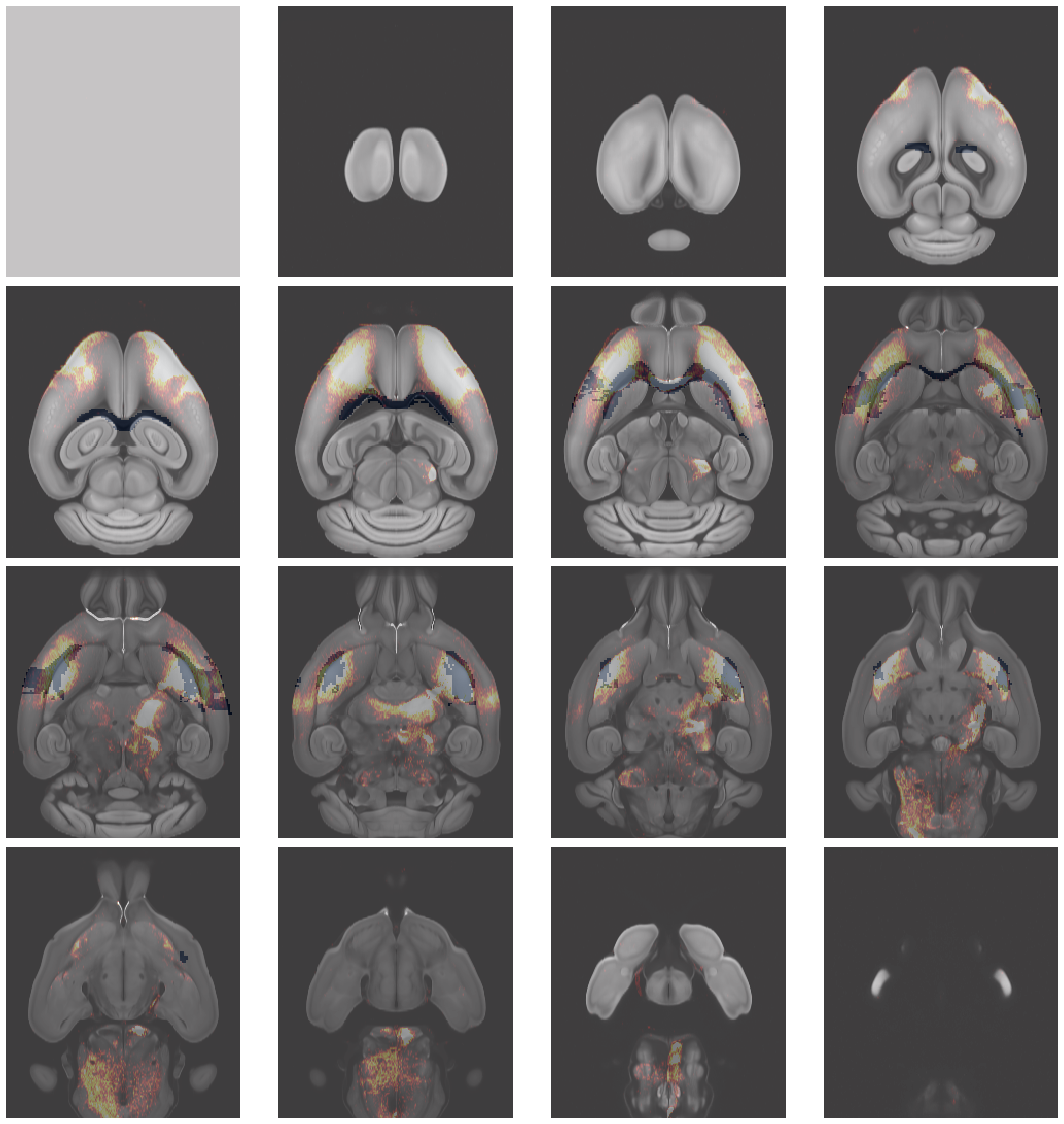
BIN
output/figures/Figure 2024-01-26 143629.png
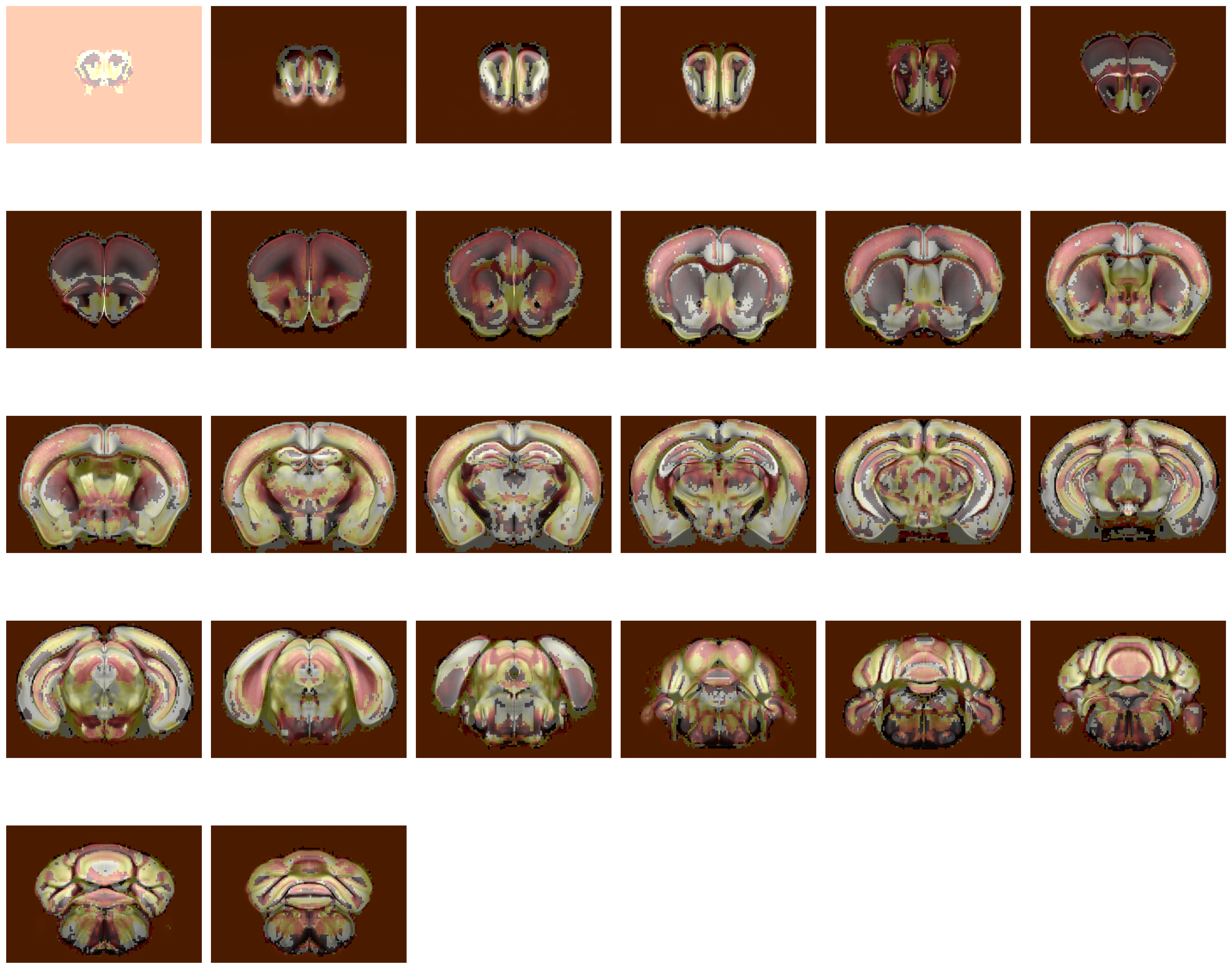
BIN
output/figures/Figure 2024-01-26 153529.png
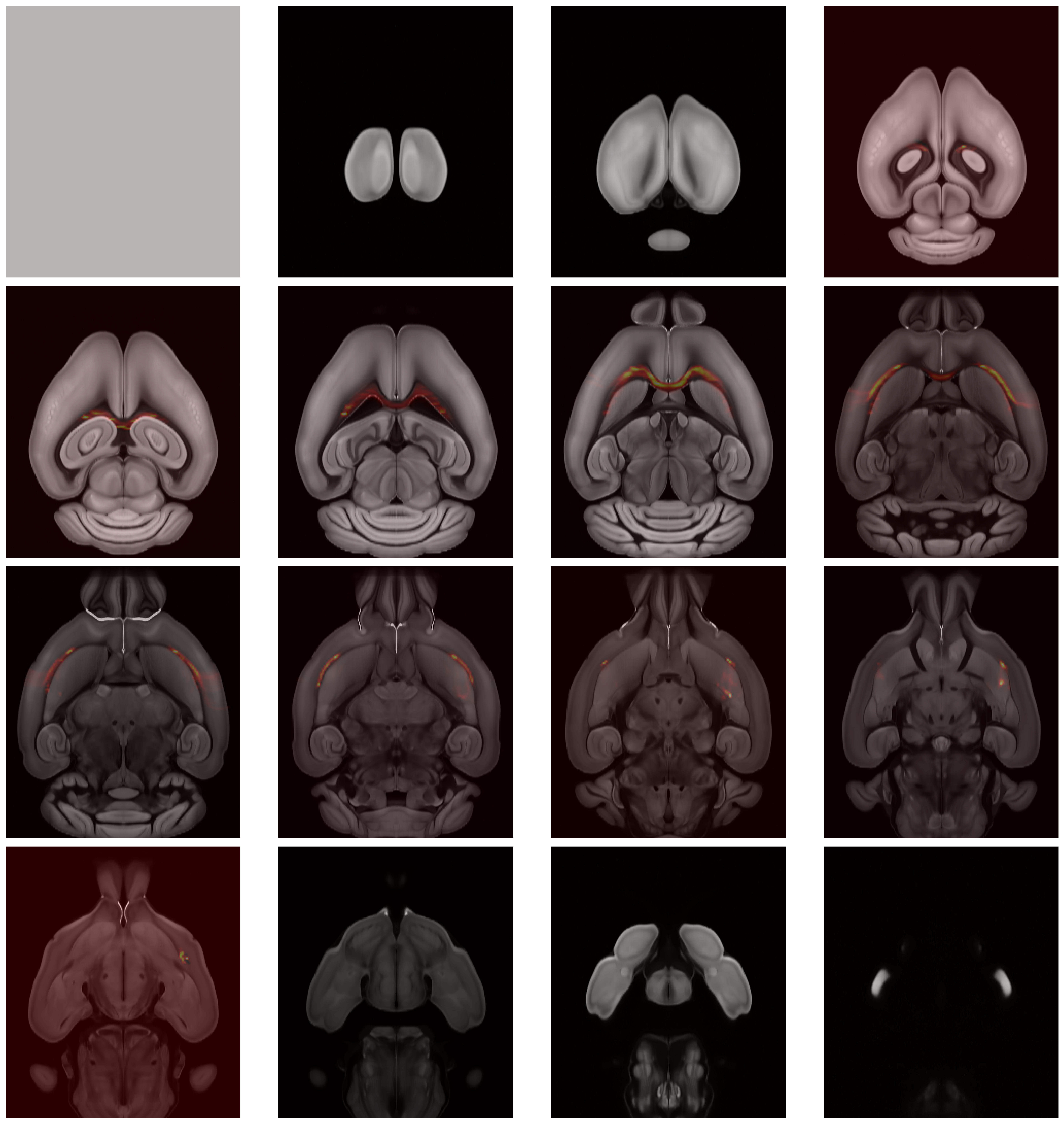
BIN
output/figures/Figure 2024-01-26 153824.png