100 mengubah file dengan 133508 tambahan dan 250239 penghapusan
+ 1
- 1
code/correlation_between_bahavior_and_DTI.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
File diff ditekan karena terlalu besar
+ 3888
- 3888
output/Correlation_with_behavior/correlation_dti_with_behavior.csv
File diff ditekan karena terlalu besar
+ 3889
- 1297
output/Correlation_with_behavior/del/correlation_dti_with_behavior.csv
File diff ditekan karena terlalu besar
+ 122472
- 122472
output/Final_Quantitative_output/Quantitative_results_from_dwi_processing.csv
File diff ditekan karena terlalu besar
+ 1
- 122473
output/Final_Quantitative_output/Quantitative_results_from_dwi_processing_merged_with_behavior_data.csv
+ 67
- 0
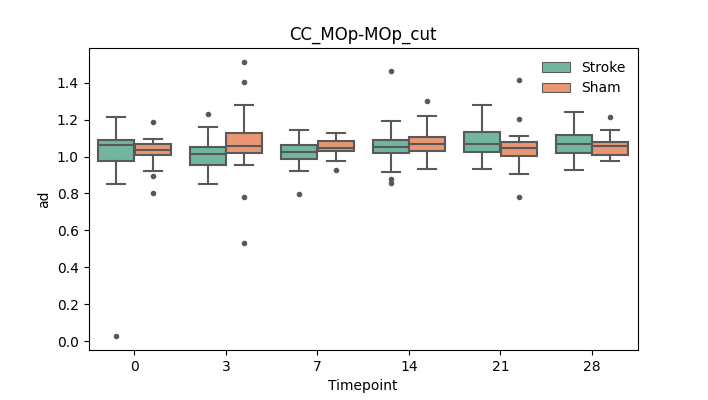
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_MOp-MOp_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_MOp-MOp_cut_mixedlm_significant.png
+ 67
- 0
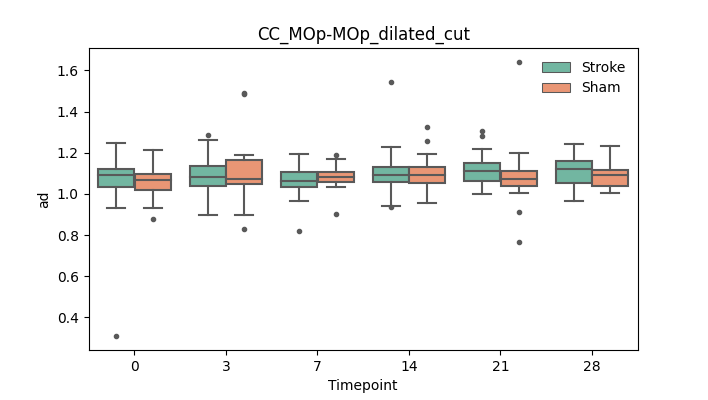
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_MOp-MOp_dilated_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_MOp-MOp_dilated_cut_mixedlm_significant.png
+ 67
- 0
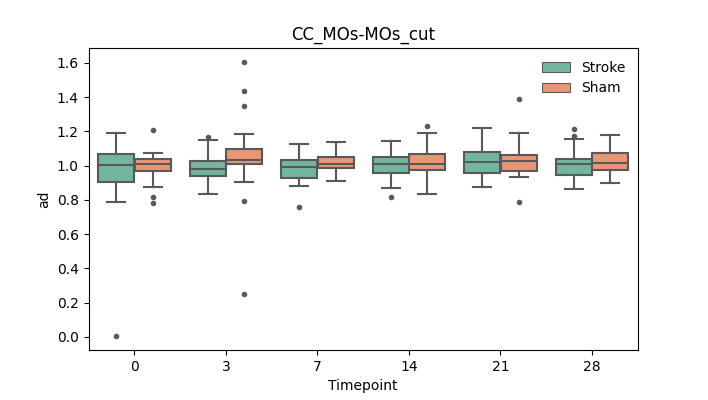
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_MOs-MOs_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_MOs-MOs_cut_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_SSp_ll-SSp_ll_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_SSp_ll-SSp_ll_cut_mixedlm_significant.png
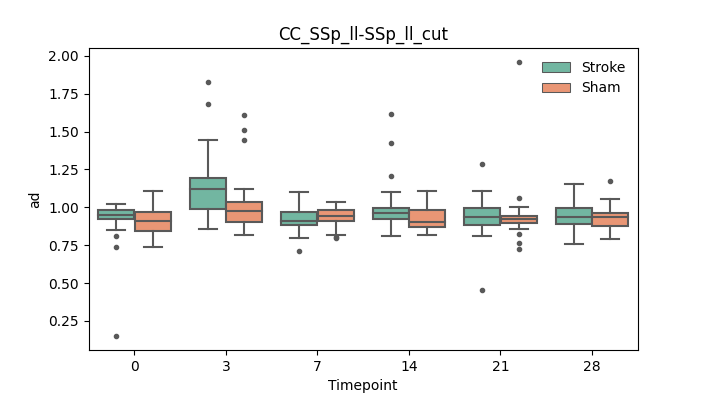
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_SSp_ul-SSp_ul_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CC_SSp_ul-SSp_ul_cut_mixedlm_significant.png
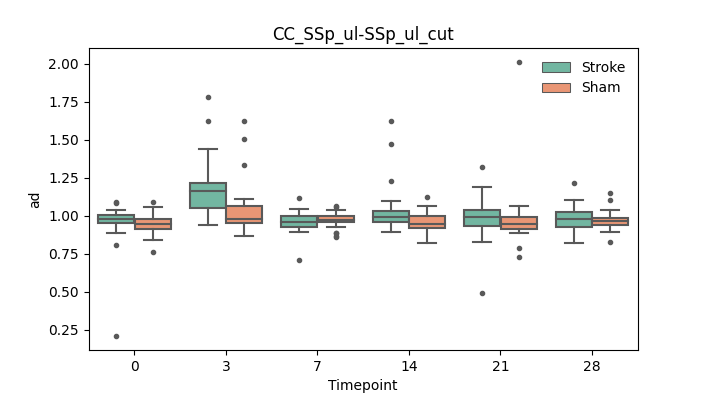
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_CReT_MOp-TRN_ipsilesional_CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CReT_MOp-TRN_ipsilesional_CCcut_mixedlm_significant.png
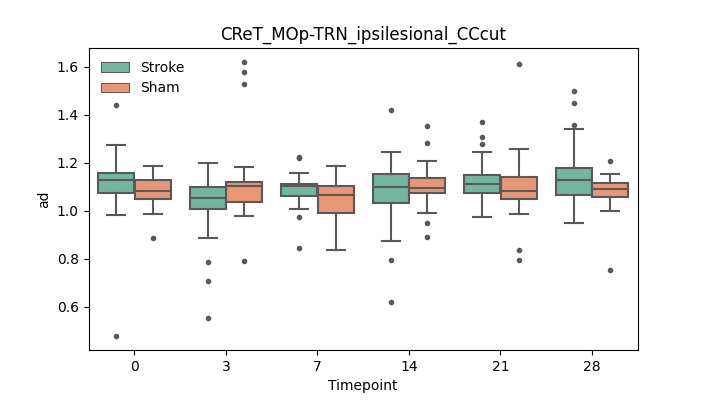
+ 67
- 0
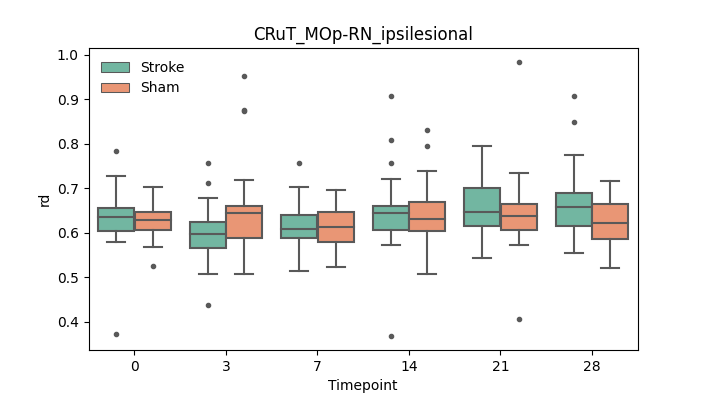
output/Final_Quantitative_output/mixed_model_analysis/ad_CRuT_MOp-RN_ipsilesional_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CRuT_MOp-RN_ipsilesional_mixedlm_significant.png
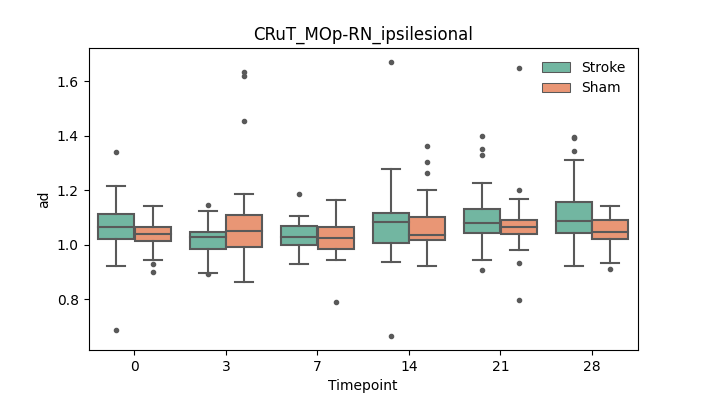
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_CST_MOp-int-py_contralesional_CCasROA_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CST_MOp-int-py_contralesional_CCasROA_mixedlm_significant.png
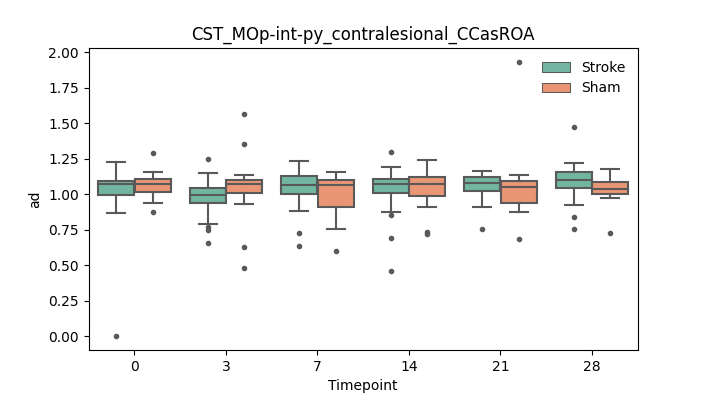
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_CST_MOp-int-py_contralesional_CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CST_MOp-int-py_contralesional_CCcut_mixedlm_significant.png
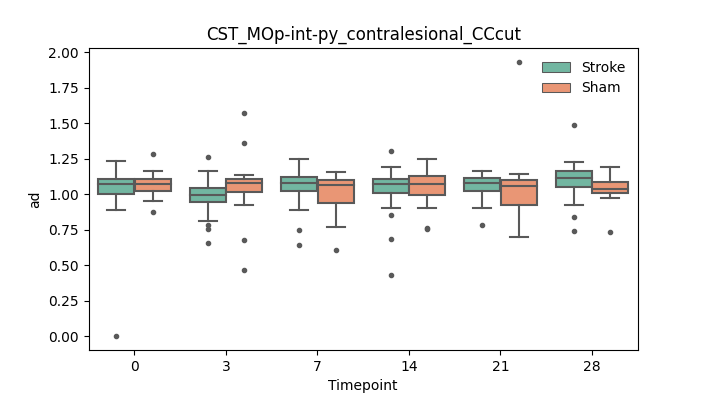
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_CST_MOp-int-py_contralesional_selfdrawnROA+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_CST_MOp-int-py_contralesional_selfdrawnROA+CCcut_mixedlm_significant.png
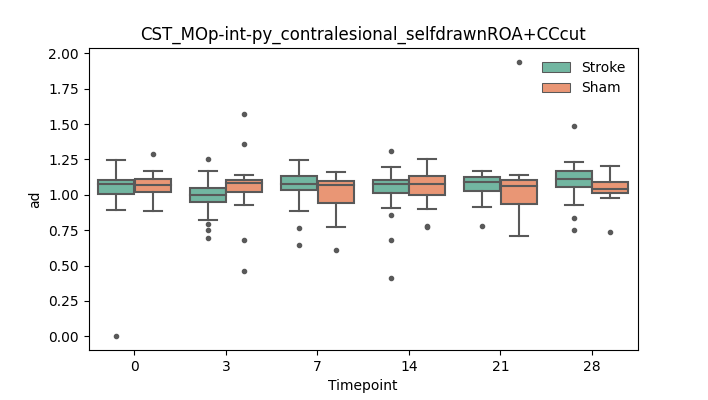
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_END+higherstepsize__log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_END+higherstepsize__mixedlm_significant.png
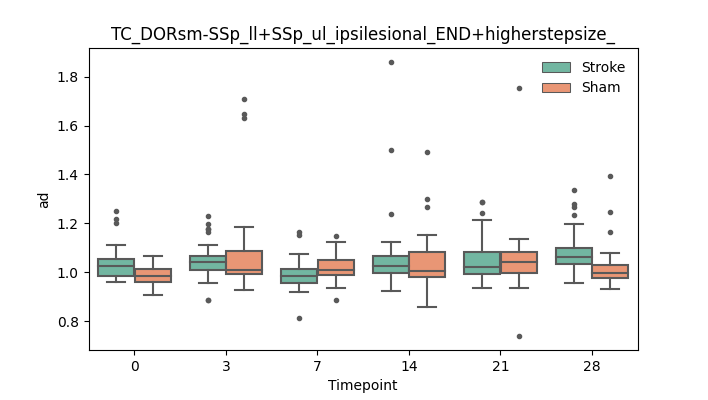
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/ad_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_ROI+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/ad_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_ROI+CCcut_mixedlm_significant.png
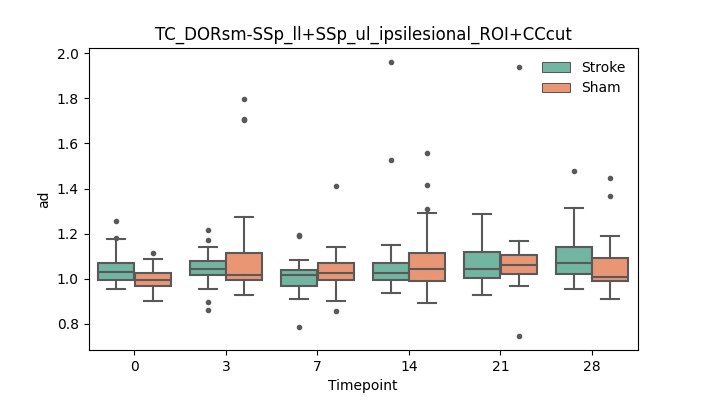
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_MOp-MOp_dilated_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_MOp-MOp_dilated_mixedlm_significant.png
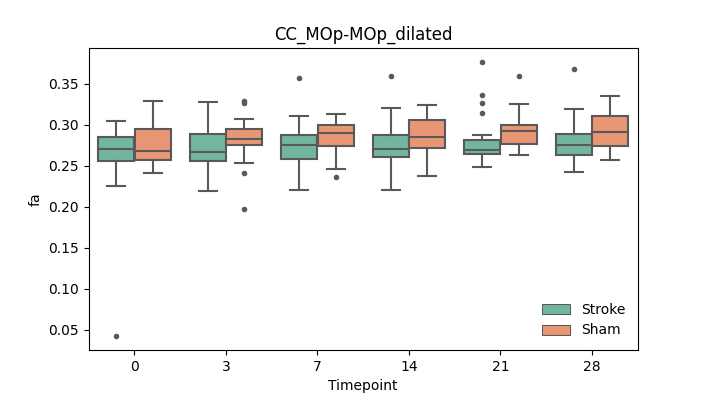
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_MOs-MOs_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_MOs-MOs_mixedlm_significant.png
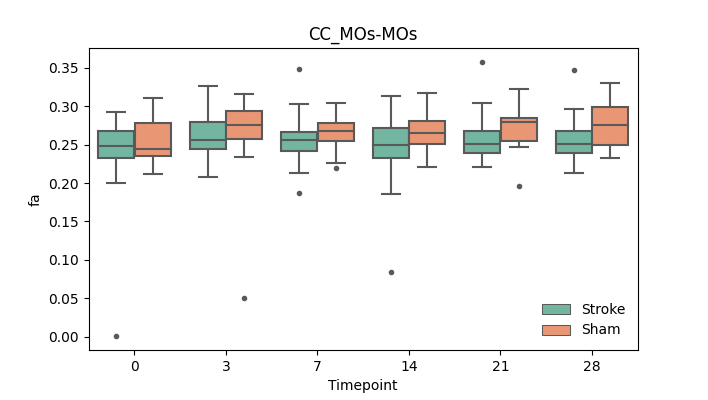
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ll-SSp_ll_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ll-SSp_ll_cut_mixedlm_significant.png
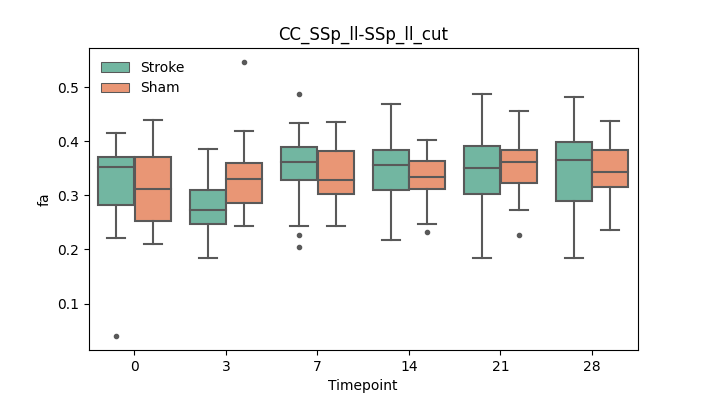
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ll-SSp_ll_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ll-SSp_ll_mixedlm_significant.png
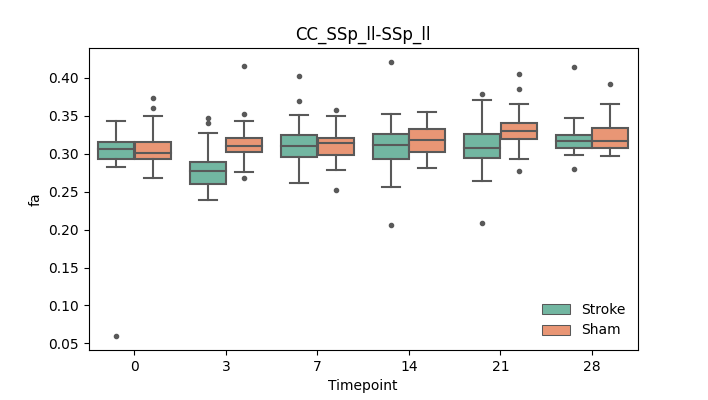
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ul-SSp_ul_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ul-SSp_ul_cut_mixedlm_significant.png
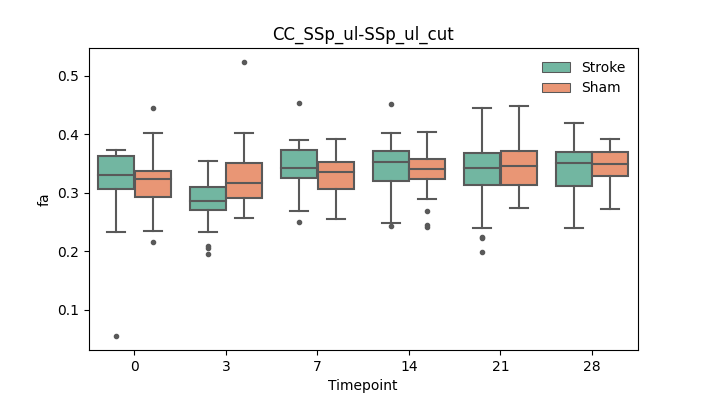
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ul-SSp_ul_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CC_SSp_ul-SSp_ul_mixedlm_significant.png
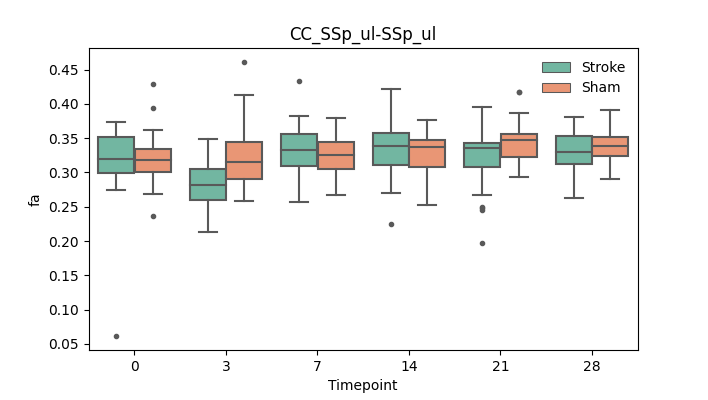
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CReT_MOp-TRN_contralesional_CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CReT_MOp-TRN_contralesional_CCcut_mixedlm_significant.png
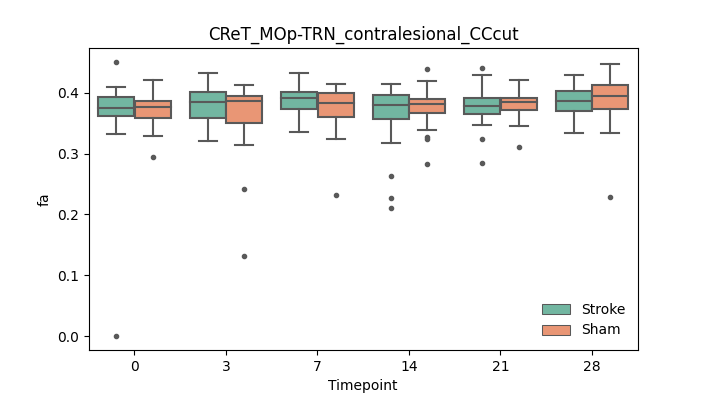
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CRuT_MOp-RN_contralesional_mirrored_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CRuT_MOp-RN_contralesional_mirrored_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_contralesional_CCasROA_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_contralesional_CCasROA_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_contralesional_CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_contralesional_CCcut_mixedlm_significant.png

+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_contralesional_selfdrawnROA+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_contralesional_selfdrawnROA+CCcut_mixedlm_significant.png

+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_ipsilesional_CCasROA_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_ipsilesional_CCasROA_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_ipsilesional_selfdrawnROA+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_CST_MOp-int-py_ipsilesional_selfdrawnROA+CCcut_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp+SSs_contralesional_END+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp+SSs_contralesional_END+CCcut_mixedlm_significant.png
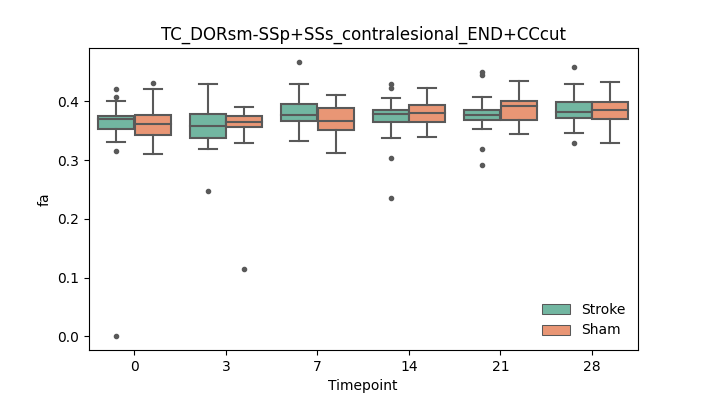
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp+SSs_ipsilesional_END+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp+SSs_ipsilesional_END+CCcut_mixedlm_significant.png
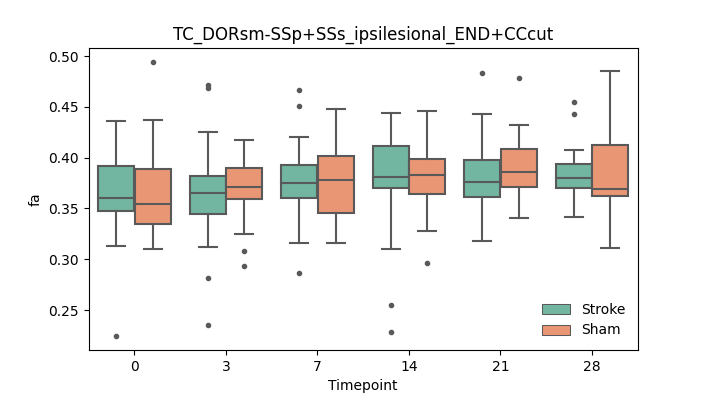
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_contralesional_END+higherstepsize_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_contralesional_END+higherstepsize_mixedlm_significant.png
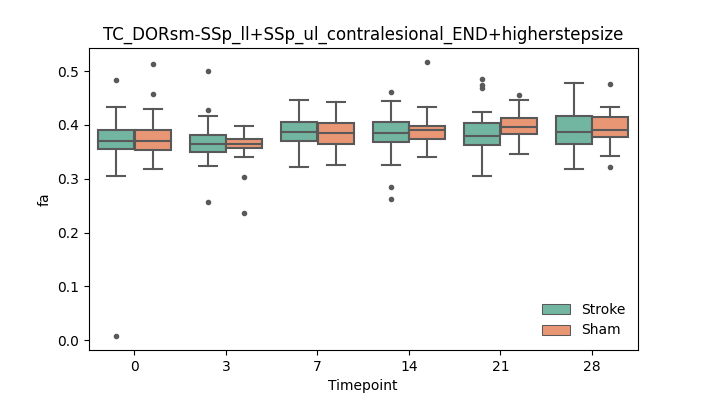
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_contralesional_ROI+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_contralesional_ROI+CCcut_mixedlm_significant.png
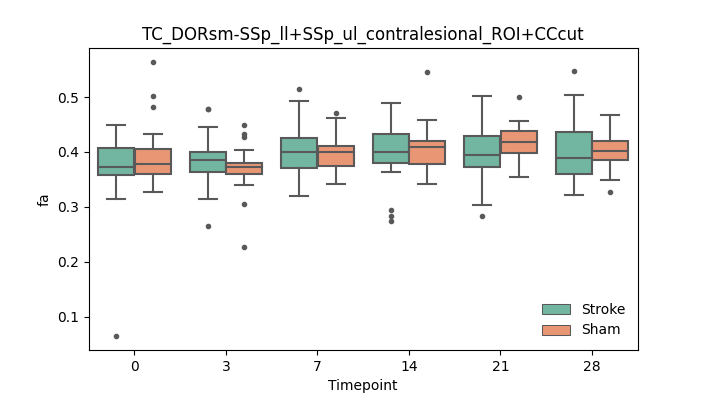
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_END+higherstepsize__log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_END+higherstepsize__mixedlm_significant.png
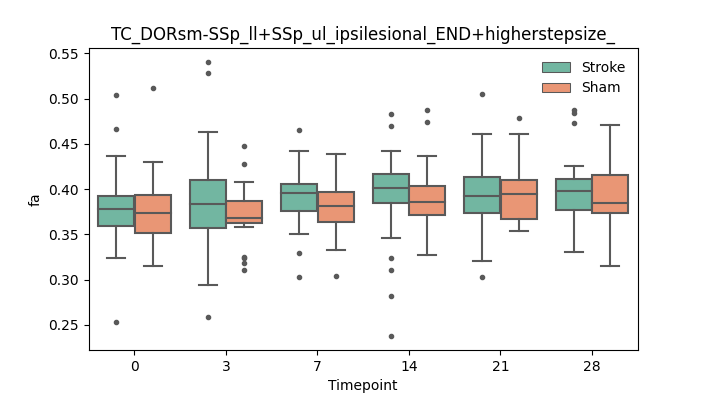
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_ROI+CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/fa_TC_DORsm-SSp_ll+SSp_ul_ipsilesional_ROI+CCcut_mixedlm_significant.png
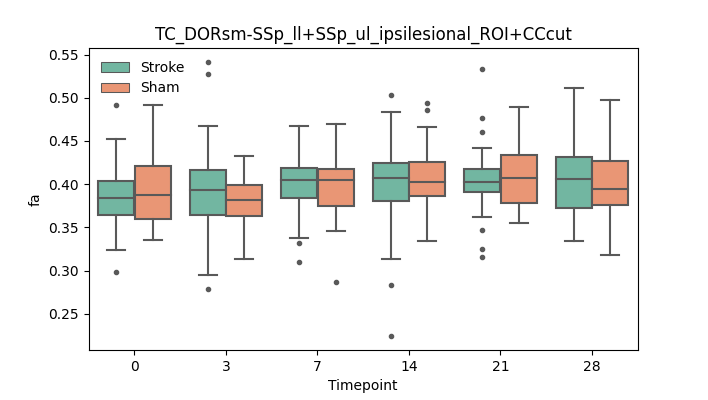
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/md_CC_MOp-MOp_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/md_CC_MOp-MOp_cut_mixedlm_significant.png
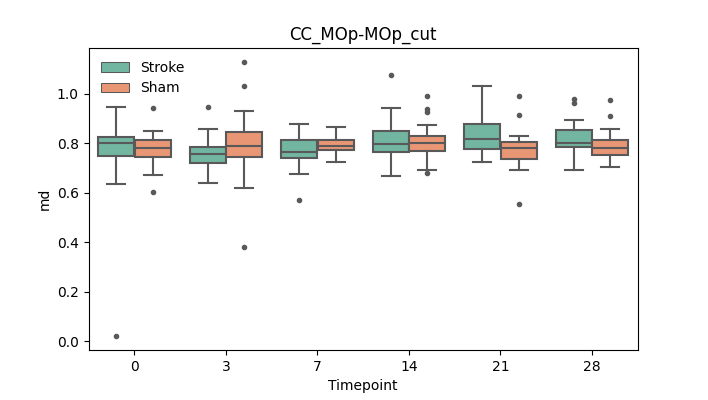
+ 67
- 0
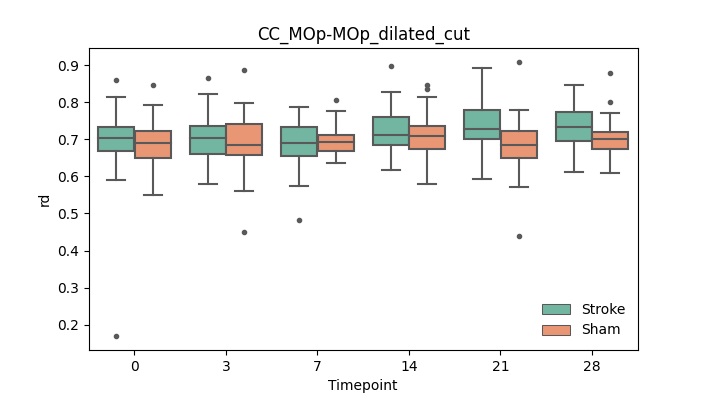
output/Final_Quantitative_output/mixed_model_analysis/md_CC_MOp-MOp_dilated_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/md_CC_MOp-MOp_dilated_cut_mixedlm_significant.png
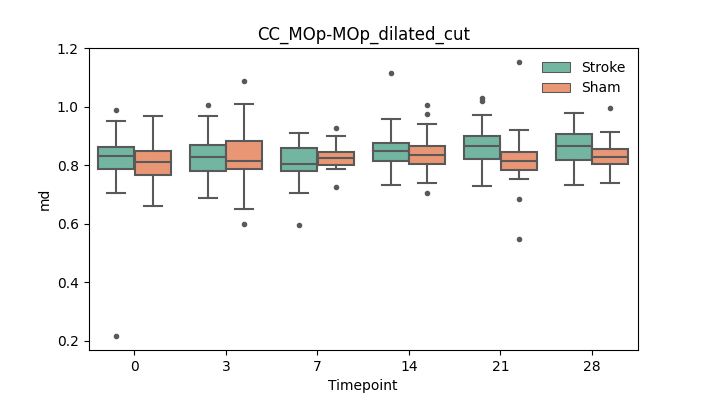
+ 67
- 0
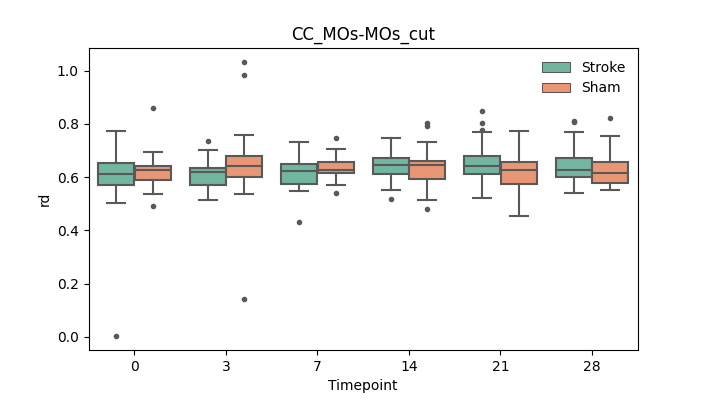
output/Final_Quantitative_output/mixed_model_analysis/md_CC_MOs-MOs_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/md_CC_MOs-MOs_cut_mixedlm_significant.png
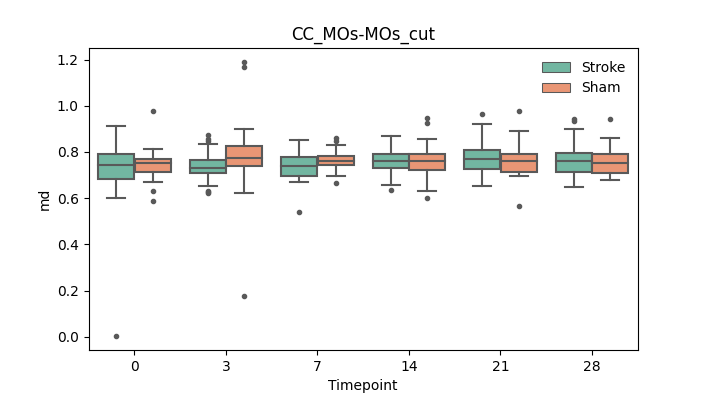
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/md_CC_SSp_ll-SSp_ll_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/md_CC_SSp_ll-SSp_ll_cut_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/md_CC_SSp_ul-SSp_ul_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
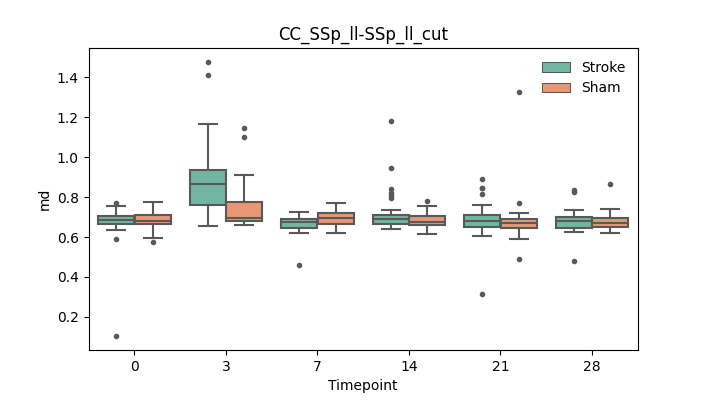
output/Final_Quantitative_output/mixed_model_analysis/md_CC_SSp_ul-SSp_ul_cut_mixedlm_significant.png
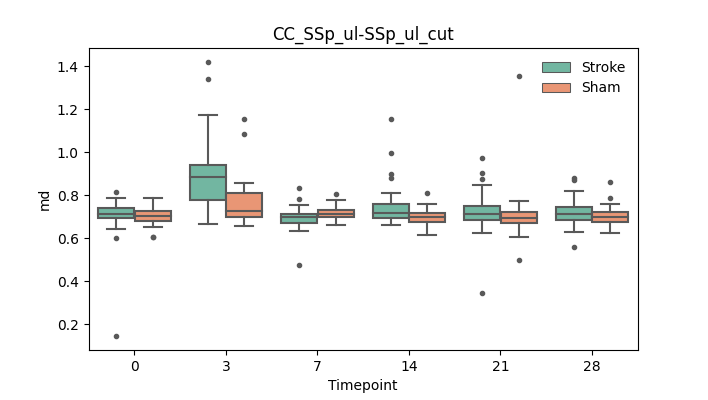
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/md_CReT_MOp-TRN_ipsilesional_CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/md_CReT_MOp-TRN_ipsilesional_CCcut_mixedlm_significant.png
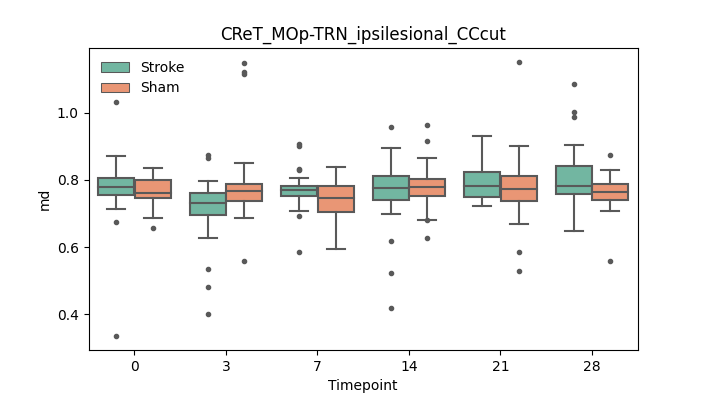
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/md_CRuT_MOp-RN_ipsilesional_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
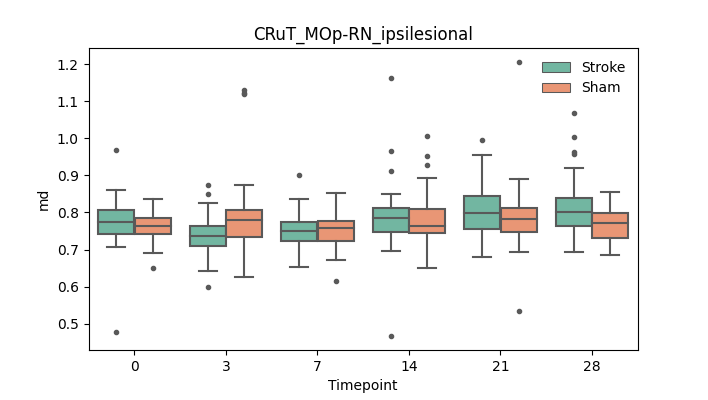
output/Final_Quantitative_output/mixed_model_analysis/md_CRuT_MOp-RN_ipsilesional_mixedlm_significant.png
+ 108
- 108
output/Final_Quantitative_output/mixed_model_analysis/mixed_model_results.csv
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 67
- 0
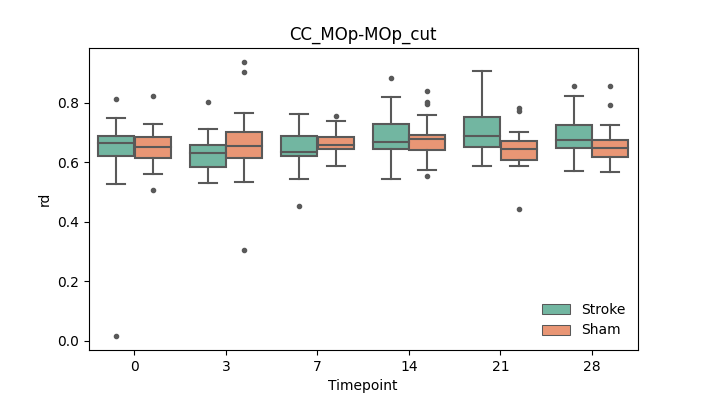
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_MOp-MOp_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_MOp-MOp_cut_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_MOp-MOp_dilated_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_MOp-MOp_dilated_cut_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_MOs-MOs_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_MOs-MOs_cut_mixedlm_significant.png
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ll-SSp_ll_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ll-SSp_ll_cut_mixedlm_significant.png
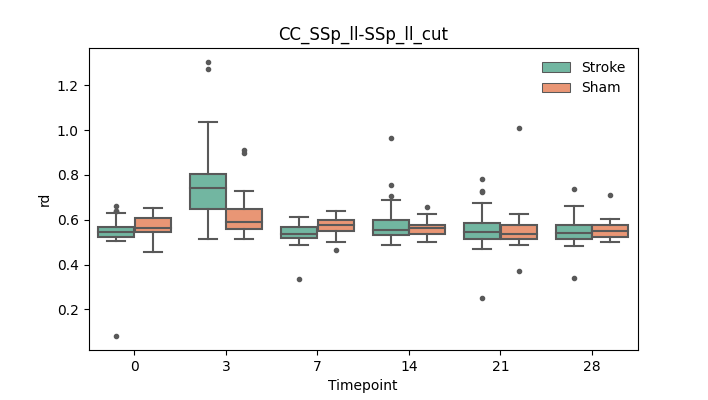
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ll-SSp_ll_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ll-SSp_ll_mixedlm_significant.png
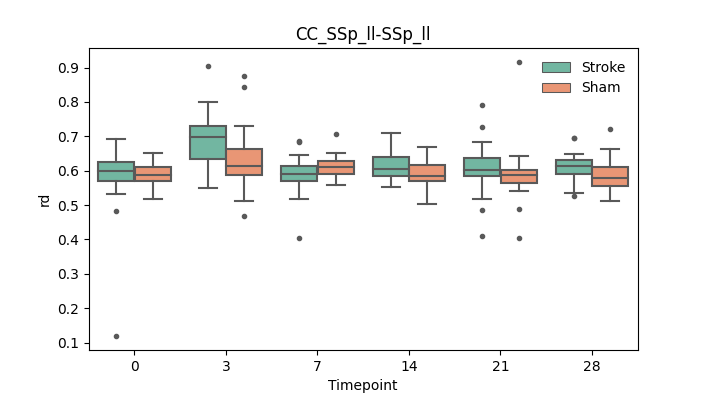
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ul-SSp_ul_cut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ul-SSp_ul_cut_mixedlm_significant.png
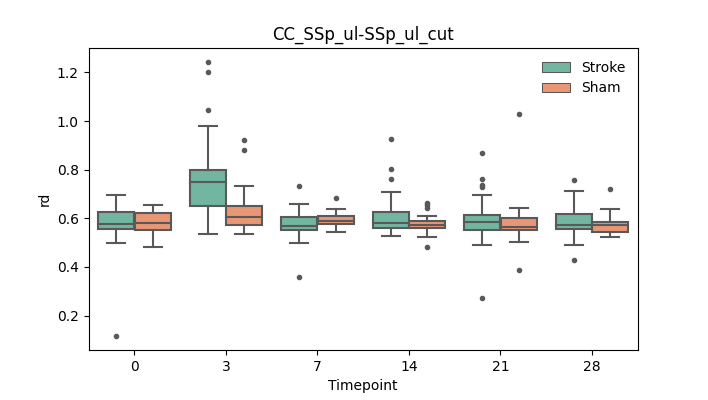
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ul-SSp_ul_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CC_SSp_ul-SSp_ul_mixedlm_significant.png
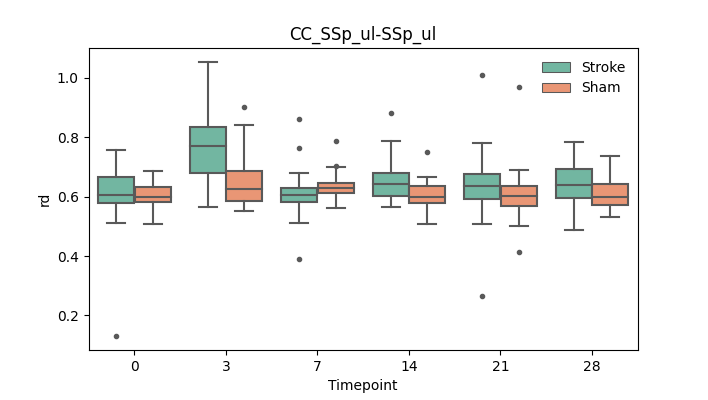
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CReT_MOp-TRN_ipsilesional_CCcut_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
output/Final_Quantitative_output/mixed_model_analysis/rd_CReT_MOp-TRN_ipsilesional_CCcut_mixedlm_significant.png
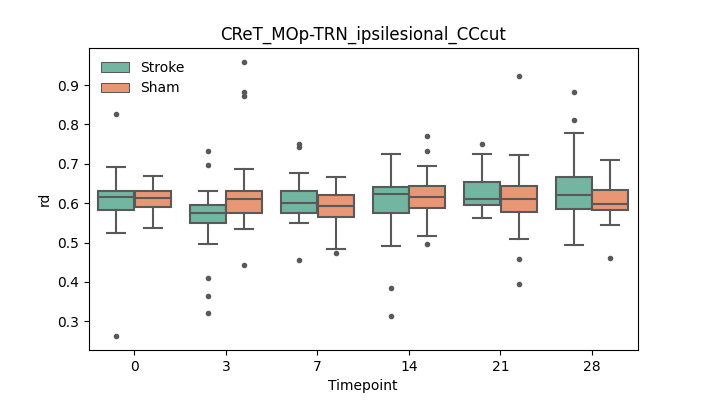
+ 67
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CRuT_MOp-RN_ipsilesional_log.txt
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 0
- 0
output/Final_Quantitative_output/mixed_model_analysis/rd_CRuT_MOp-RN_ipsilesional_mixedlm_significant.png