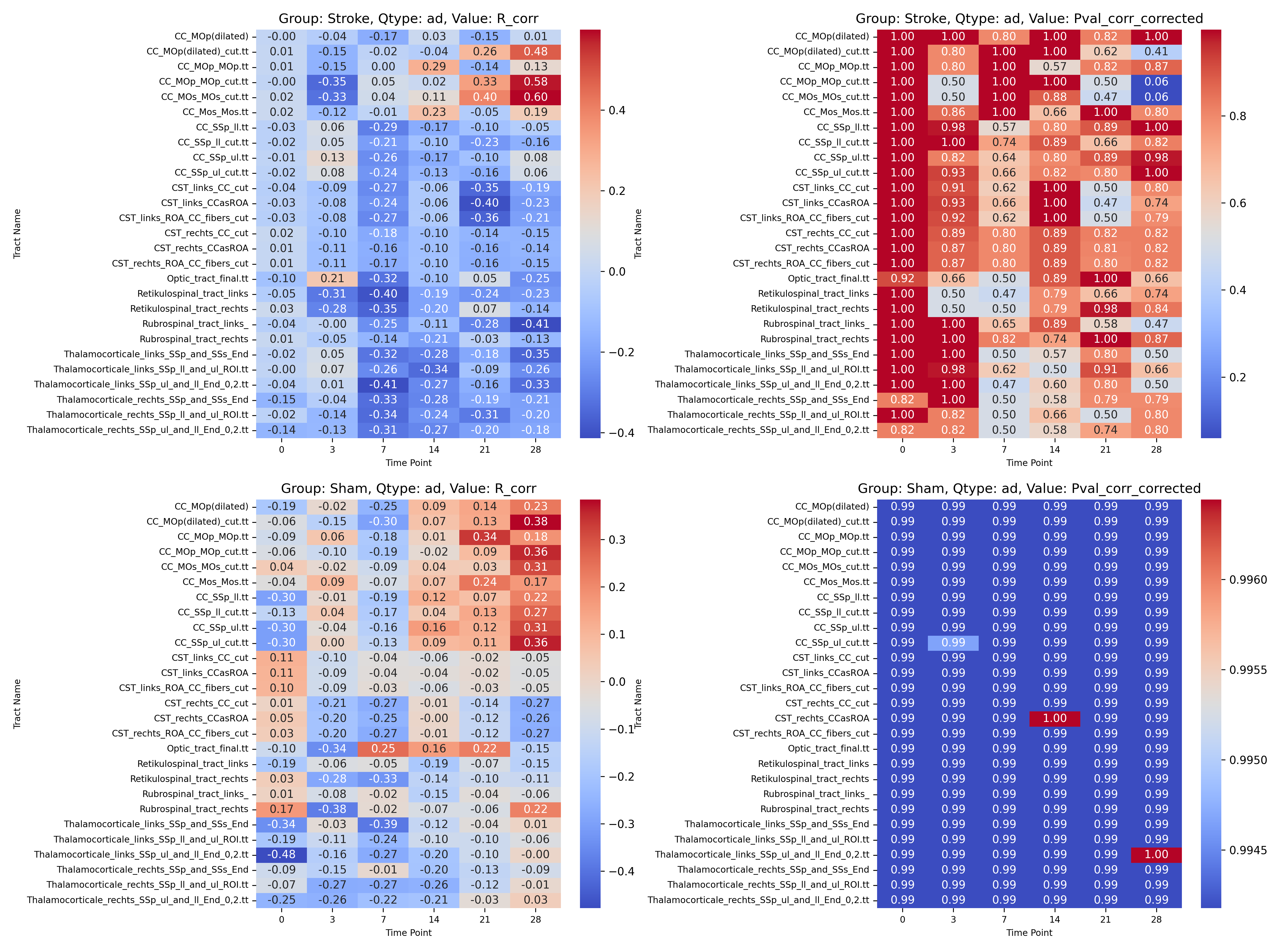
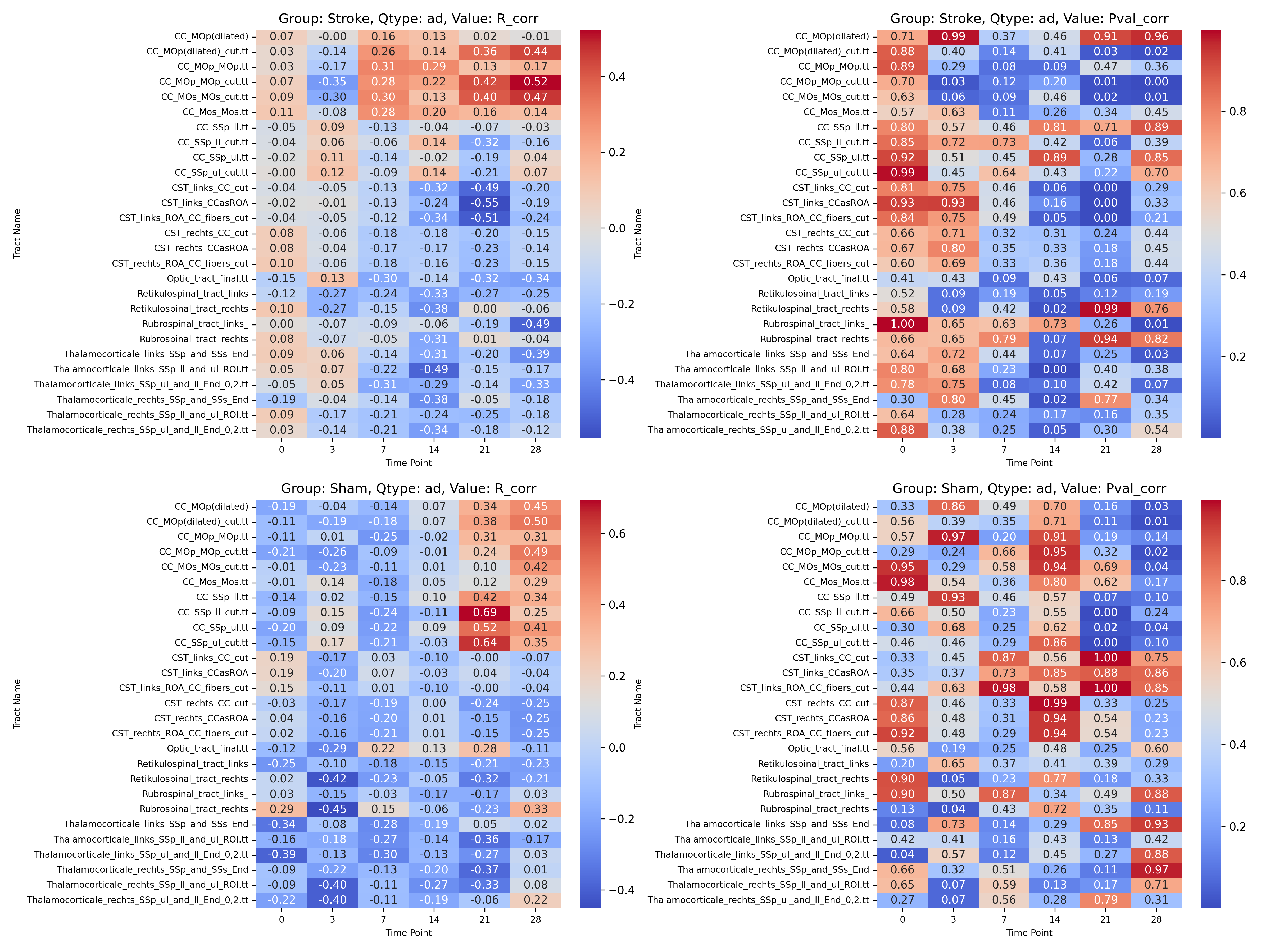
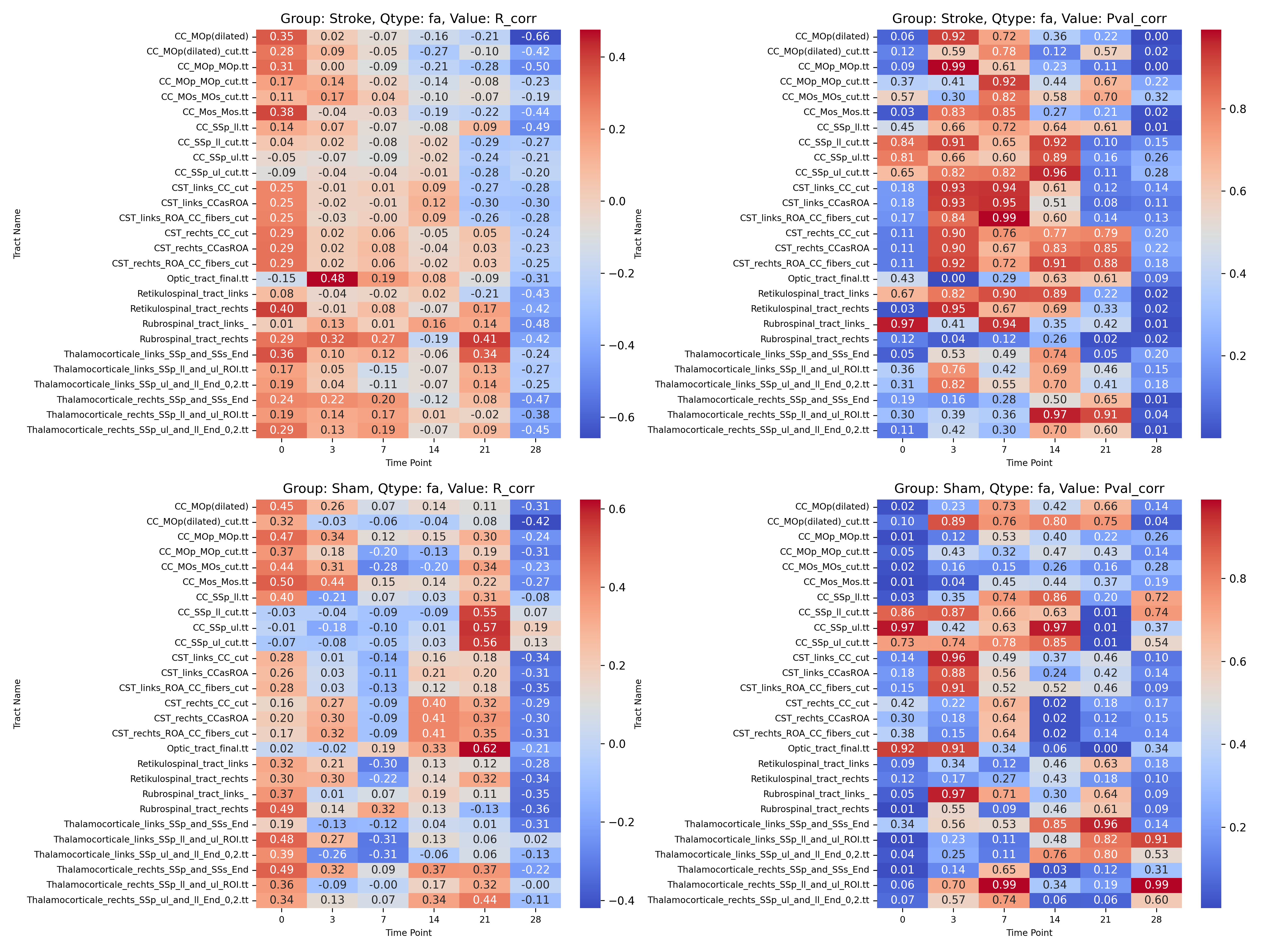
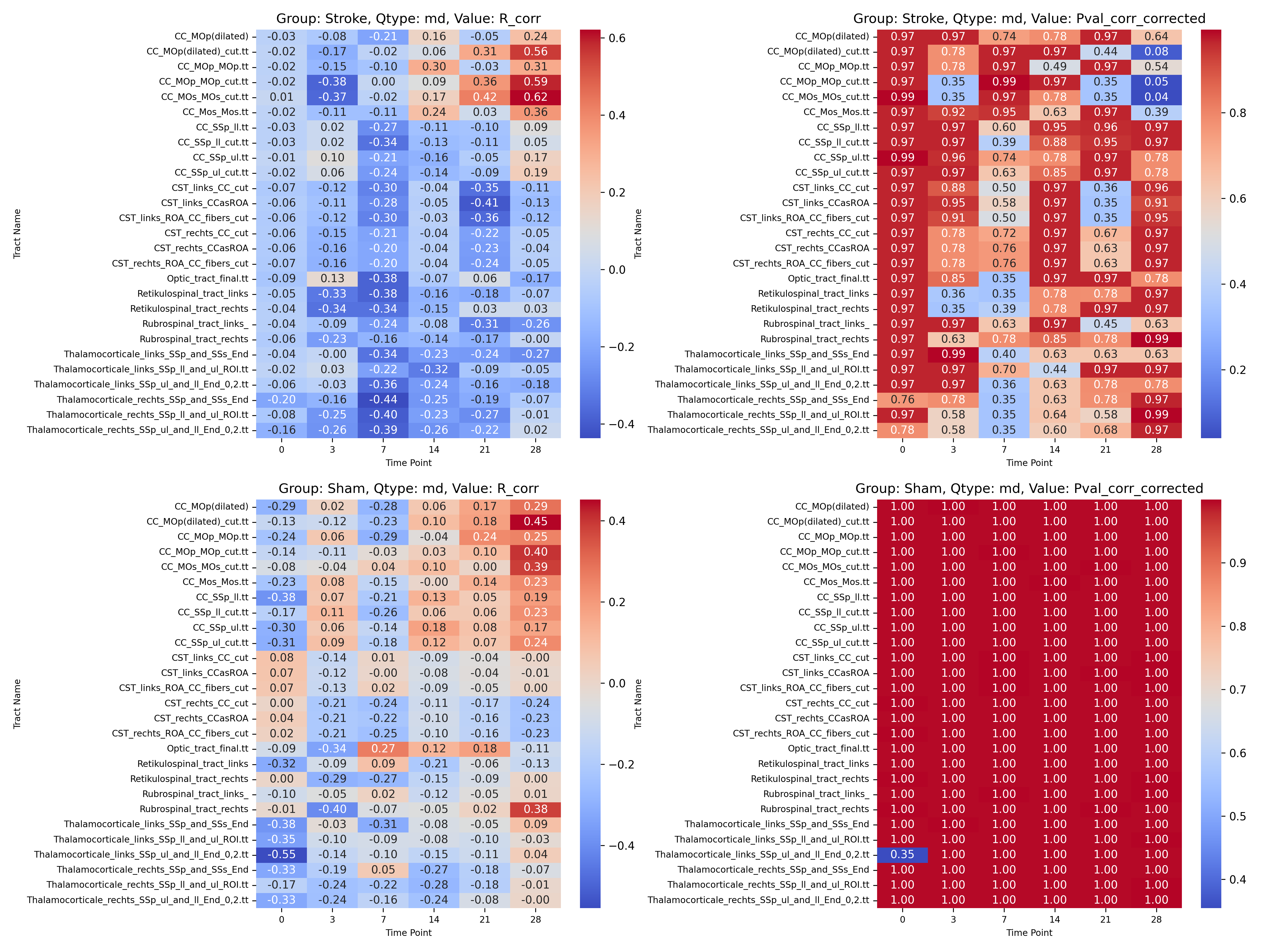
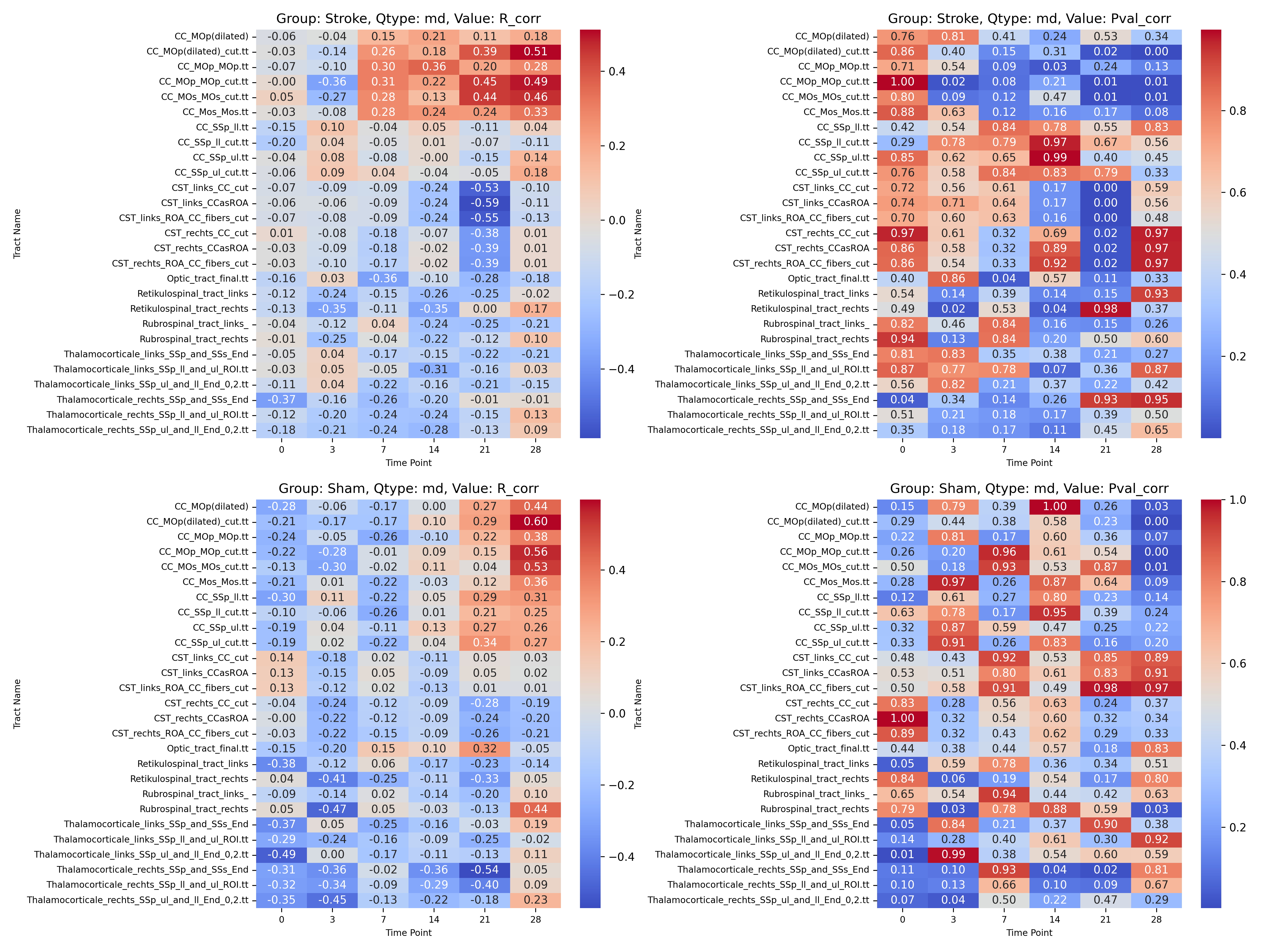
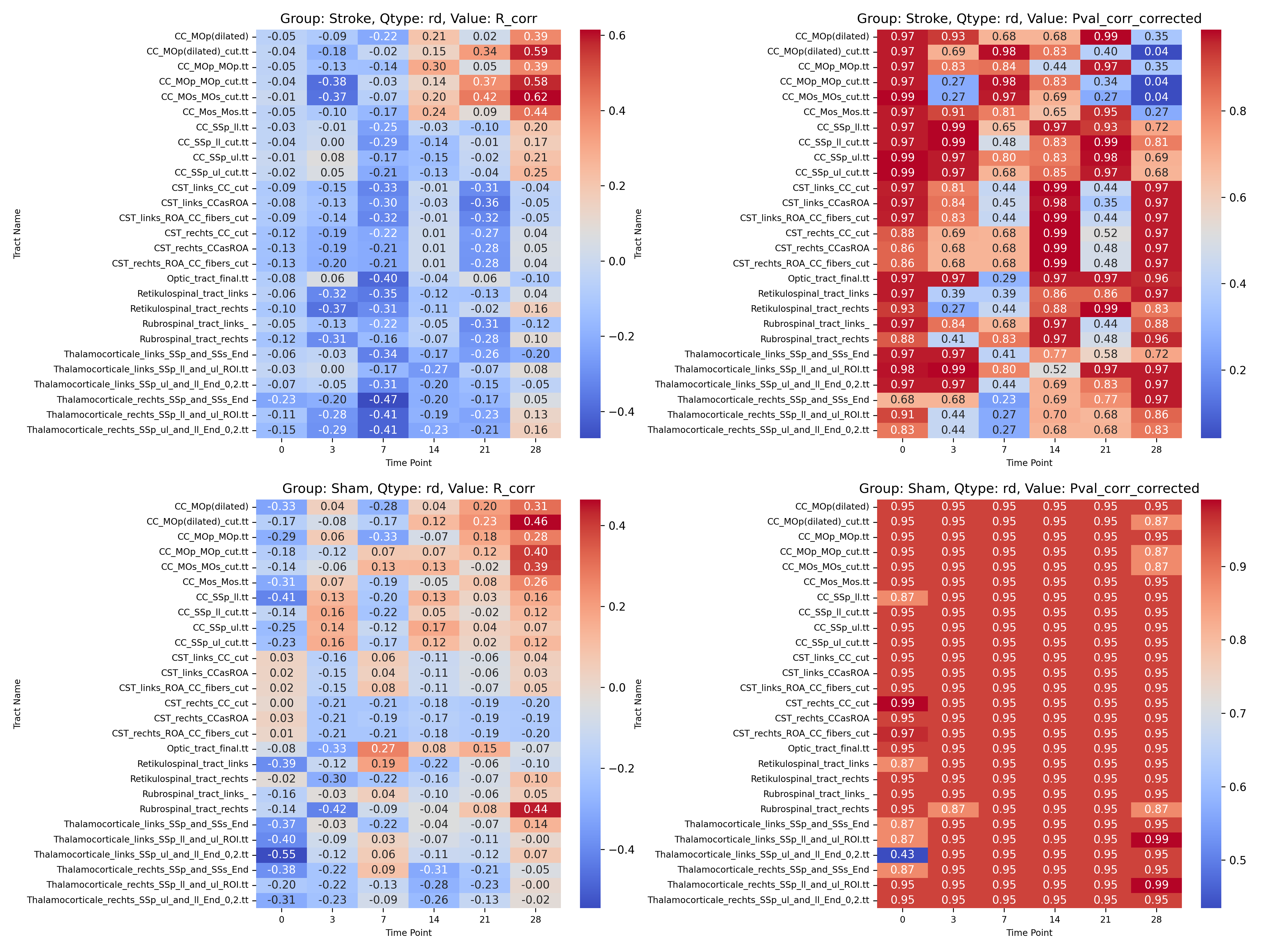
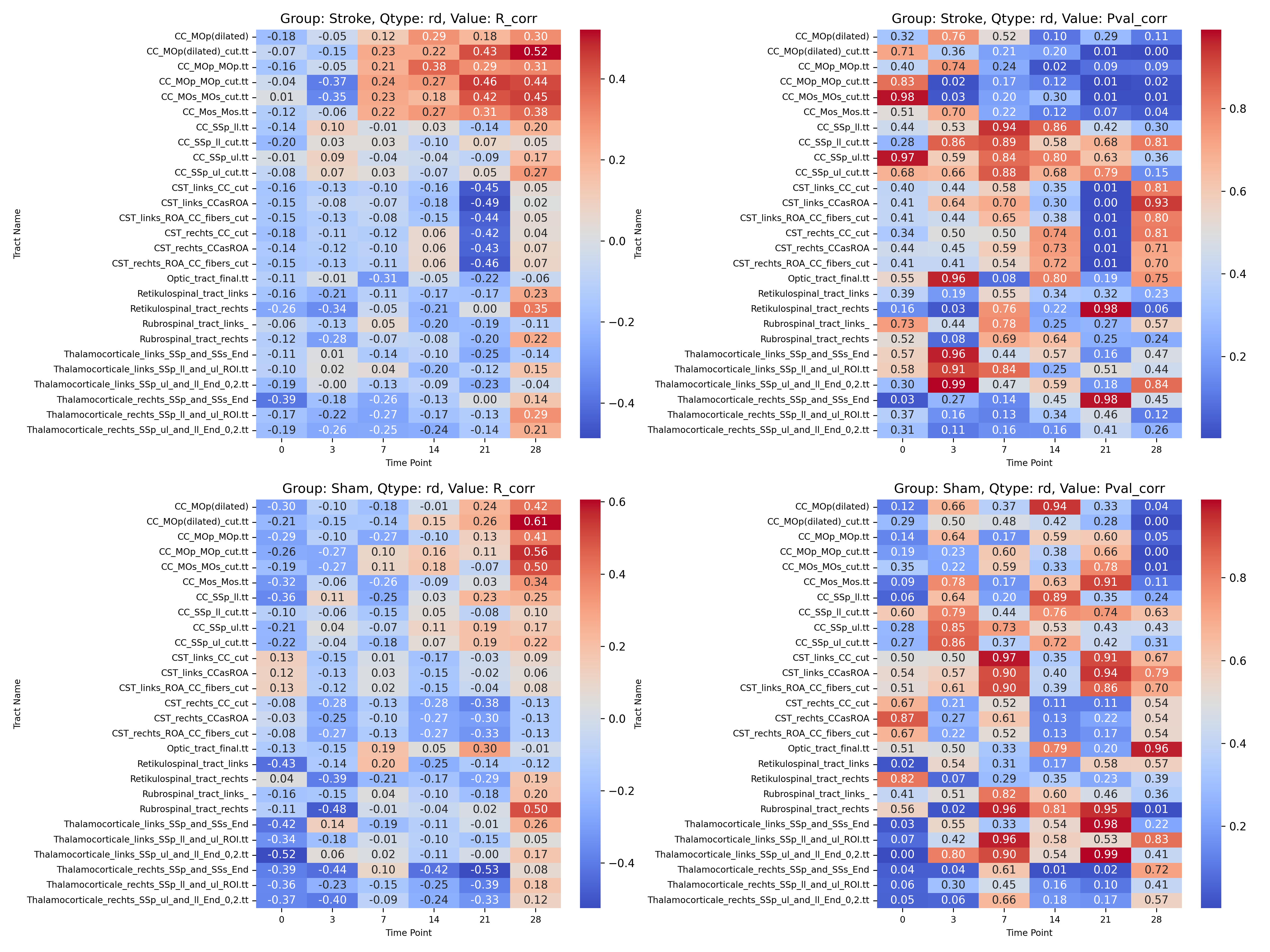
|
|
@@ -1,7 +1,8 @@
|
|
|
import os
|
|
|
import pandas as pd
|
|
|
-from scipy.stats import pearsonr
|
|
|
+from scipy.stats import spearmanr, shapiro
|
|
|
import numpy as np
|
|
|
+from statsmodels.stats.multitest import multipletests
|
|
|
|
|
|
# Get the directory where the code file is located
|
|
|
code_dir = os.path.dirname(os.path.abspath(__file__))
|
|
|
@@ -10,11 +11,12 @@ code_dir = os.path.dirname(os.path.abspath(__file__))
|
|
|
parent_dir = os.path.dirname(code_dir)
|
|
|
|
|
|
# Step 4: Save the resulting dataframe to a CSV file
|
|
|
-input_file_path = os.path.join(parent_dir, 'output', "Final_Quantitative_output",'Final_Quantitative_output&behavior_data.csv')
|
|
|
+input_file_path = os.path.join(parent_dir, 'output', "Final_Quantitative_output", 'Quantitative_results_from_dwi_processing_merged_with_behavior_data.csv')
|
|
|
|
|
|
df = pd.read_csv(input_file_path)
|
|
|
|
|
|
-df_f1 = (df["dialation_amount"] == 0)
|
|
|
+
|
|
|
+df_f1 = (df["dialation_amount"] == 2)
|
|
|
|
|
|
# Empty list to store the results
|
|
|
results = []
|
|
|
@@ -26,42 +28,49 @@ for ss in df["Group"].unique():
|
|
|
for mm in df["mask_name"].unique():
|
|
|
# Filter the DataFrame for the current combination of 'Group', 'merged_timepoint', 'Qtype', and 'mask_name'
|
|
|
df_f2 = df[df_f1 & (df["Group"] == ss) & (df["merged_timepoint"] == time_point) & (df["Qtype"] == qq) & (df["mask_name"] == mm)]
|
|
|
-
|
|
|
+
|
|
|
# Remove rows with NaN values in 'Value' or 'averageScore' columns
|
|
|
df_f2 = df_f2.dropna(subset=['Value', 'averageScore'])
|
|
|
-
|
|
|
+
|
|
|
if not df_f2.empty:
|
|
|
- # Calculate Pearson correlation coefficient between 'Value' and 'averageScore' columns
|
|
|
- correlation_coefficient, p_value = pearsonr(df_f2["Value"], df_f2["averageScore"])
|
|
|
+
|
|
|
+ correlation_coefficient, p_value = spearmanr(df_f2["Value"], df_f2["averageScore"])
|
|
|
+ # Calculate Shapiro-Wilk test for normality
|
|
|
+ shapiro_statValue, shapiro_pvalueQValue = shapiro(df_f2["Value"])
|
|
|
+ shapiro_statScore, shapiro_pvalueBehavior = shapiro(df_f2["averageScore"])
|
|
|
|
|
|
+
|
|
|
# Store the results in a dictionary
|
|
|
- result = {'Group': ss, 'merged_timepoint': time_point, 'Qtype': qq, 'mask_name': mm, 'Pval_corr': p_value, 'R_corr': correlation_coefficient}
|
|
|
-
|
|
|
+ result = {'Group': ss, 'merged_timepoint': time_point, 'Qtype': qq, 'mask_name': mm,
|
|
|
+ 'Pval_corr': p_value, 'R_corr': correlation_coefficient,
|
|
|
+ 'shapiro-wilk_pvalue_qtype': shapiro_pvalueQValue,'shapiro-wilk_pvalue_behavior': shapiro_pvalueBehavior}
|
|
|
+
|
|
|
# Append the dictionary to the results list
|
|
|
results.append(result)
|
|
|
else:
|
|
|
- print(f"No valid data found for Group: {ss}, merged_timepoint: {time_point}, Qtype: {qq}, and mask_name: {mm}. Skipping.")
|
|
|
+ print(
|
|
|
+ f"No valid data found for Group: {ss}, merged_timepoint: {time_point}, Qtype: {qq}, and mask_name: {mm}. Skipping.")
|
|
|
|
|
|
# Create a DataFrame from the results list
|
|
|
correlation_results_df = pd.DataFrame(results)
|
|
|
|
|
|
-# Apply Holm-Bonferroni correction to p-values
|
|
|
-p_values = correlation_results_df['Pval_corr']
|
|
|
-num_tests = len(p_values)
|
|
|
-sorted_indices = np.argsort(p_values)
|
|
|
-
|
|
|
-# Initialize an empty array to store the corrected p-values
|
|
|
-p_values_corrected = np.ones(num_tests)
|
|
|
-
|
|
|
-# Loop through the sorted p-values and apply the Holm-Bonferroni correction
|
|
|
-for i, idx in enumerate(sorted_indices):
|
|
|
- p_values_corrected[idx] = np.min([1, p_values[idx] * (num_tests - i)])
|
|
|
+# Apply FDR correction to p-values within each Qtype and each Group separately
|
|
|
+unique_qtypes = correlation_results_df['Qtype'].unique()
|
|
|
+unique_groups = df["Group"].unique()
|
|
|
+unique_masks = df["mask_name"].unique()
|
|
|
|
|
|
-# Assign the corrected p-values to the DataFrame
|
|
|
-correlation_results_df['Pval_corr_corrected'] = p_values_corrected
|
|
|
+for qtype in unique_qtypes:
|
|
|
+ for mask in unique_masks:
|
|
|
+ mask_mask = correlation_results_df['mask_name'] == mask
|
|
|
+ qtype_group_mask = mask_mask & (correlation_results_df['Qtype'] == qtype)
|
|
|
+ p_values = correlation_results_df[qtype_group_mask]['Pval_corr']
|
|
|
+ rejected, p_values_corrected, _, _ = multipletests(p_values, method='fdr_bh')
|
|
|
|
|
|
+ # Assign the corrected p-values to the DataFrame
|
|
|
+ correlation_results_df.loc[qtype_group_mask, 'Pval_corr_corrected'] = p_values_corrected
|
|
|
+
|
|
|
# Define the output file path
|
|
|
-output_file_path = os.path.join(parent_dir, 'output', "Correlation_with_behavior",'correlation_results.csv')
|
|
|
+output_file_path = os.path.join(parent_dir, 'output', "Correlation_with_behavior", 'correlation_dti_with_behavior5.csv')
|
|
|
|
|
|
# Save the correlation results DataFrame to a CSV file
|
|
|
correlation_results_df.to_csv(output_file_path, index=False)
|