Gallant Lab Natural Short Clips 3T fMRI Data
Summary
This dataset contains BOLD fMRI responses in human subjects viewing a set of
natural short movie clips. The functional data were collected in five subjects,
in three sessions over three separate days for each subject. Details of the
experiment are described in the original publication [1].
[1] Huth, Alexander G., Nishimoto, S., Vu, A. T., & Gallant, J. L.
(2012). A continuous semantic space describes the representation of thousands
of object and action categories across the human brain. Neuron, 76(6),
1210-1224. https://dx.doi.org/10.1016/j.neuron.2012.10.014
Cite this dataset
If you publish any work using the dataset, please cite the original publication
[1], and cite the dataset [1b] in the following recommended format:
[1b] Huth, A. G., Nishimoto, S., Vu, A. T., Dupre la Tour, T., & Gallant,
J. L. (2022). Gallant Lab Natural Short Clips 3T fMRI Data.
https://dx.doi.org/--TBD--
Difference with the "vim-2" dataset
The present dataset uses the same stimuli (natural short movie clips) than a
previous experiment of the Gallant lab [2], publicly released in CRCNS under
the name "vim-2" [2b]. Both dataset
use the same stimuli, but the functional data is different.
The "shortclips" dataset [1b] contains full brain responses recorded every two
seconds (2s) with a 3T scanner. The "vim-2" dataset [2b] contains responses
from the occipital lobe only, recorded every second (1s) with a 4T scanner.
Contrary to the "shortclips" dataset, the "vim-2" dataset does not provide
mappers to plot the data on flatten maps of the cortical surface.
[2] Nishimoto, S., Vu, A. T., Naselaris, T., Benjamini, Y., Yu, B., &
Gallant, J. L. (2011). Reconstructing visual experiences from brain activity
evoked by natural movies. Current Biology, 21(19), 1641-1646.
https://dx.doi.org/10.1016/j.cub.2011.08.031
[2b] Nishimoto, S., Vu, A. T., Naselaris, T., Benjamini, Y., Yu, B., &
Gallant, J. L. (2014). Gallant Lab Natural Movie 4T fMRI Data. CRCNS.org.
https://dx.doi.org/10.6080/K00Z715X
How to get started
a. With dedicated tutorials
The preferred way to explore this dataset is through the voxelwise
tutorials. These tutorials
includes Python downloading tools, data loaders, plotting utilities, and
examples of analysis following the original publication [1] [2].
To run the tutorials, see
https://gallantlab.github.io/voxelwise_tutorials).
b. With git and git-annex
To download the data with git-annex, the
following dependencies are necessary: git, git-annex. Then, run the commands
# clone the repository, without the data files
git clone https://gin.g-node.org/gallantlab/shortclips
cd shortclips
# download one file (e.g. features/wordnet.hdf)
git annex get features/wordnet.hdf --from wasabi
# download all files
git annex get . --from wasabi
To maximize the download speed, two remotes are available to download the data.
The first remote is GIN (--from origin), but the bandwidth might be limited.
The second remote is Wasabi (--from wasabi), with a larger bandwidth.
A basic example script is available in example.py. For more utilities and
example of analysis, see the dedicated voxelwise
tutorials.
How to get help
The recommended way to ask questions is in the issue tracker on the GitHub page
https://github.com/gallantlab/voxelwise_tutorials/issues.
Data files organization
features/ → feature spaces used for voxelwise modeling
motion_energy.hdf → visual motion energy, as described in [2]
wordnet.hdf → visual semantic labels, as described in [1]
mappers/ → plotting mappers for each subject
S01_mapper.hdf
...
S05_mapper.hdf
responses/ → functional responses for each subject
S01_responses.hdf
...
S05_responses.hdf
stimuli/ → natural movie stimuli, for each fMRI run
test.hdf
train_00.hdf
...
train_11.hdf
utils/
wordnet_categories.txt → names of the wordnet labels
wordnet_graph.dot → wordnet graph to plot as in [1]
example.py → Python example to load and plot the data
Data format
All files are hdf5 files, with multiple arrays stored inside.
The names, shapes, and descriptions of each array are listed below.
Each file in `features` contains:
X_train: array of shape (3600, n_features)
Training features.
X_test: array of shape (270, n_features)
Testing features.
run_onsets: array of shape (12, )
Indices of each run onset.
where (n_features = 6555) for `motion_energy.hdf`
and (n_features = 1705) for `wordnet.hdf`.
Each file in `mappers` contains:
voxel_to_flatmap: CSR sparse array of shape (n_pixels, n_voxels)
Mapper from voxels to flatmap image. The sparse array is stored with
four dense arrays: (data, indices, indptr, shape).
voxel_to_fsaverage: CSR sparse array of shape (n_vertices, n_voxels)
Mapper from voxels to FreeSurfer surface. The sparse array is stored
with four dense arrays: (data, indices, indptr, shape).
flatmap_mask: array of shape (width, height)
Pixels of the flatmap image associated with a voxel.
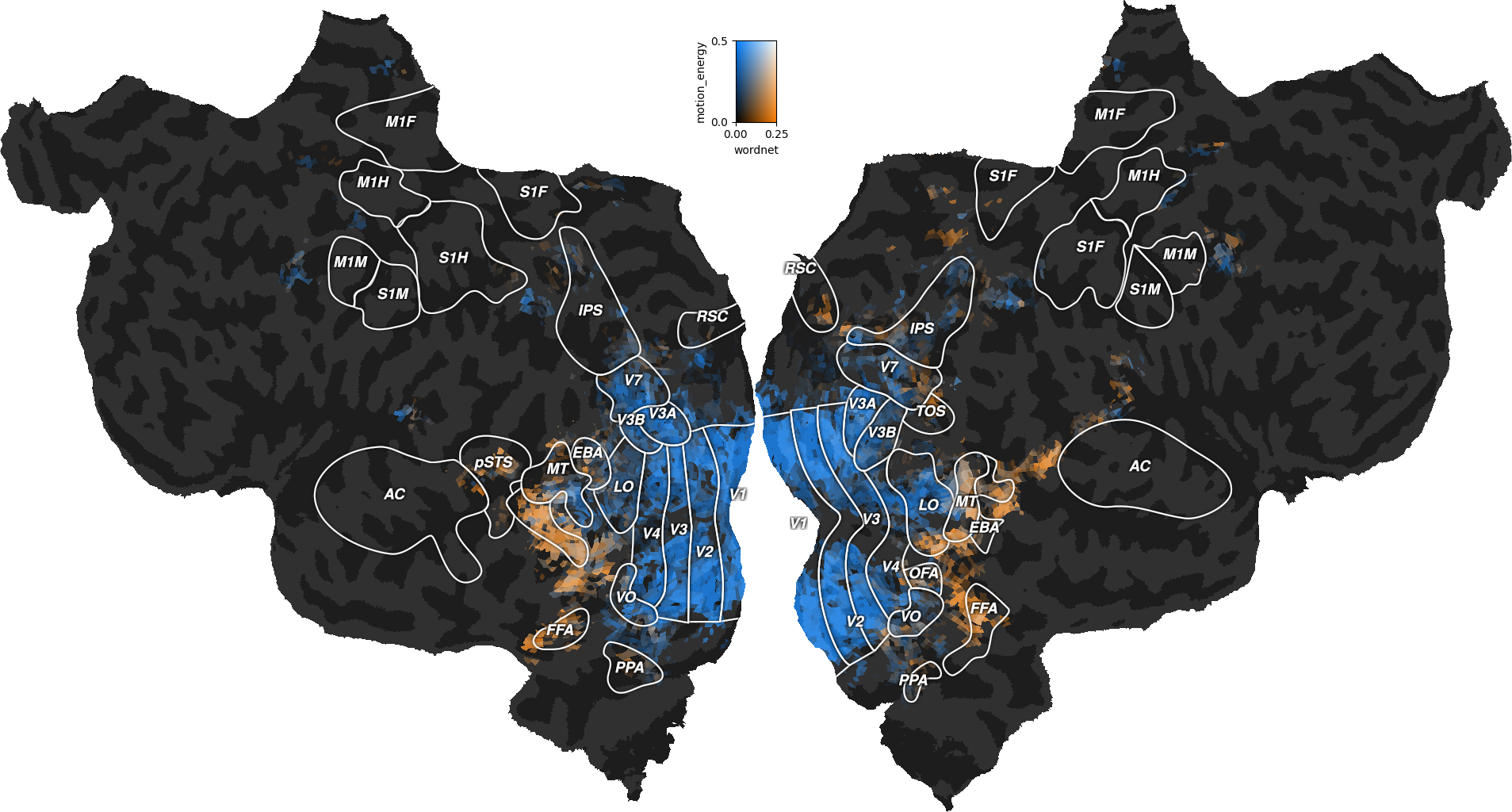
flatmap_rois: array of shape (width, height, 4)
Transparent image with annotated ROIs (for subjects S01, S02, and S03).
flatmap_curvature: array of shape (width, height)
Transparent image with binarized curvature to locate sulci/gyri.
roi_mask_xxx: array of shape (n_voxels, )
Mask indicating which voxels are in the ROI `xxx`.
ROI list is different on each subject. SO4 and S05 have no ROIs.
Each file in `responses` contains:
Y_train: array of shape (3600, n_voxels)
Training responses.
Y_test: array of shape (270, n_voxels)
Testing responses.
run_onsets: array of shape (12, )
Indices of each run onset.
Each file in `stimuli` contains:
stimuli: array of shape (n_images, 512, 512, 3)
Each training run contains 9000 images total.
The test run contains 8100 images total.
