|
|
@@ -0,0 +1,457 @@
|
|
|
+#!/usr/bin/env python
|
|
|
+# coding: utf-8
|
|
|
+
|
|
|
+# ### Define paths and animals for the analysis
|
|
|
+
|
|
|
+# In[1]:
|
|
|
+
|
|
|
+
|
|
|
+path = '/media/andrey/My Passport/GIN/Anesthesia_CA1/meta_data/meta_recordings_transition_state.xlsx'
|
|
|
+path4results = '/media/andrey/My Passport/GIN/Anesthesia_CA1/validation/calcium_imaging_transition_state/' #To store transformation matrix
|
|
|
+save_plots_path = '/media/andrey/My Passport/GIN/Anesthesia_CA1/validation/calcium_imaging_transition_state/'
|
|
|
+log_file_path = save_plots_path + 'registration_logs.txt'
|
|
|
+
|
|
|
+animals_for_analysis = [8237]
|
|
|
+
|
|
|
+# ### Align FOV's for all recordings
|
|
|
+
|
|
|
+# In[2]:
|
|
|
+
|
|
|
+
|
|
|
+repeat_calc = 1
|
|
|
+silent_mode = False
|
|
|
+
|
|
|
+#######################
|
|
|
+
|
|
|
+import pandas as pd
|
|
|
+import numpy as np
|
|
|
+import os
|
|
|
+
|
|
|
+import matplotlib.pyplot as plt
|
|
|
+
|
|
|
+import sys
|
|
|
+np.set_printoptions(threshold=sys.maxsize)
|
|
|
+
|
|
|
+from pystackreg import StackReg
|
|
|
+
|
|
|
+# Sobel filter (not used)
|
|
|
+#from scipy import ndimage #https://docs.scipy.org/doc/scipy/reference/generated/scipy.ndimage.sobel.html
|
|
|
+
|
|
|
+meta_data = pd.read_excel(path)
|
|
|
+
|
|
|
+#%% compute transformations matrices between recordings
|
|
|
+
|
|
|
+recordings = meta_data['Number']
|
|
|
+
|
|
|
+animals = animals_for_analysis
|
|
|
+
|
|
|
+#print("Recordings: ", recordings)
|
|
|
+
|
|
|
+#ALIGNMENT ALGORITHM
|
|
|
+
|
|
|
+# log file
|
|
|
+f = open(log_file_path, "a")
|
|
|
+
|
|
|
+print("n, i, j, rigid_mean_enh, rigid_mean, affine_mean_enh, affine_mean, best_method ", file=f)
|
|
|
+
|
|
|
+if (silent_mode!=True):
|
|
|
+ print(" RMSD's: (rigid, mean_enh) | (rigid, mean) | (affine, mean_enh) | (affine, mean) | best method ")
|
|
|
+
|
|
|
+for animal in animals:
|
|
|
+
|
|
|
+ if (silent_mode!=True):
|
|
|
+ print("Animal #", str(animal))
|
|
|
+
|
|
|
+
|
|
|
+ if not os.path.exists(path4results + 'StackReg/' +
|
|
|
+ str(animal) + '.npy') or repeat_calc == 1:
|
|
|
+# if not os.path.exists('Q:/Personal/Mattia/Calcium Imaging/results/StackRegEnhImage/' +
|
|
|
+# str(animal) + '.npy') or repeat_calc == 1:
|
|
|
+# if not os.path.exists('Q:/Personal/Mattia/Calcium Imaging/results/StackReg/' +
|
|
|
+# str(animal) + '.npy') or repeat_calc == 1:
|
|
|
+
|
|
|
+ meta_animal = meta_data[meta_data['Mouse'] == animal]
|
|
|
+ recordings = meta_animal['Number']
|
|
|
+
|
|
|
+ images_mean = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+ images_mean_enh = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+
|
|
|
+ images_quality_check = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+
|
|
|
+ best_tmats = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0], 3, 3))
|
|
|
+ best_score = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0]))
|
|
|
+ best_methods = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0]))
|
|
|
+
|
|
|
+
|
|
|
+ tmats_affine = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0], 3, 3))
|
|
|
+ tmats_rigid = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0], 3, 3))
|
|
|
+ tmats_affine_enh = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0], 3, 3))
|
|
|
+ tmats_rigid_enh = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0], 3, 3))
|
|
|
+
|
|
|
+ # load all (enhanced) images
|
|
|
+ for idx, recording in enumerate(recordings):
|
|
|
+ print(recording)
|
|
|
+ options = np.load(str(meta_data['Folder'][recording]) +
|
|
|
+ str(meta_data['Subfolder'][recording]) +
|
|
|
+ # str(int(meta_data['Recording idx'][recording])) +
|
|
|
+ '/suite2p/plane0/ops.npy',
|
|
|
+ allow_pickle=True)
|
|
|
+ # mean image or mean enhanced image
|
|
|
+ images_mean[:, :, idx] = options.item(0)['meanImg']
|
|
|
+ images_mean_enh[:, :, idx] = options.item(0)['meanImgE']
|
|
|
+ #cut_boarders=50
|
|
|
+ #quality check
|
|
|
+ images_quality_check[:, :, idx] = options.item(0)['meanImg']
|
|
|
+
|
|
|
+
|
|
|
+ # loop through every pair and compute the transformation matrix
|
|
|
+
|
|
|
+ #conditions = [meta_data['Condition'][recording] for recording in recordings]
|
|
|
+
|
|
|
+ for idx0 in range(np.shape(images_mean)[2]):
|
|
|
+ #if (idx0!=14):
|
|
|
+ #continue
|
|
|
+
|
|
|
+ for idx1 in range(idx0, np.shape(images_mean)[2]):
|
|
|
+ #if (idx1!=16):
|
|
|
+ #continue
|
|
|
+
|
|
|
+
|
|
|
+ fraction_of_non_zero_pixels = [0.0,0.0,0.0,0.0]
|
|
|
+
|
|
|
+### MEAN RIGID and AFFINE
|
|
|
+
|
|
|
+ reference_image = images_mean[:, :, idx0]
|
|
|
+ initial_image = images_mean[:, :, idx1]
|
|
|
+
|
|
|
+ #sx = ndimage.sobel(reference_image, axis=0, mode='constant')
|
|
|
+ #sy = ndimage.sobel(reference_image, axis=1, mode='constant')
|
|
|
+ #reference_image = np.hypot(sx, sy)
|
|
|
+
|
|
|
+ #sx = ndimage.sobel(initial_image, axis=0, mode='constant')
|
|
|
+ #sy = ndimage.sobel(initial_image, axis=1, mode='constant')
|
|
|
+ #initial_image = np.hypot(sx, sy)
|
|
|
+
|
|
|
+ boarder_cut = 100
|
|
|
+ sr = StackReg(StackReg.AFFINE)
|
|
|
+ tmats_affine[idx0, idx1, :, :] = sr.register(reference_image, initial_image)
|
|
|
+
|
|
|
+ image_transformed = sr.transform(images_quality_check[:, :, idx1], tmats_affine[idx0, idx1, :, :])
|
|
|
+ image_difference = images_quality_check[:, :, idx0] - image_transformed
|
|
|
+ fraction_of_non_zero_pixels[3] = np.count_nonzero(image_transformed[:,:]<0.001)/262144
|
|
|
+ #plt.imshow(image_transformed)
|
|
|
+ #plt.show()
|
|
|
+ image_difference = image_difference[boarder_cut:-boarder_cut, boarder_cut:-boarder_cut]
|
|
|
+ image_difference = np.square(image_difference)
|
|
|
+ rmsd_affine = np.sqrt(image_difference.sum()/(512 - 2 * boarder_cut)**2)
|
|
|
+ if (silent_mode!=True):
|
|
|
+ print("Fraction of non-zero pixels in 3 (mean affine): ", fraction_of_non_zero_pixels[3]," Score:",rmsd_affine)
|
|
|
+
|
|
|
+ sr = StackReg(StackReg.RIGID_BODY)
|
|
|
+ tmats_rigid[idx0, idx1, :, :] = sr.register(reference_image, initial_image)
|
|
|
+ image_transformed = sr.transform(images_quality_check[:, :, idx1], tmats_rigid[idx0, idx1, :, :])
|
|
|
+ image_difference = images_quality_check[:, :, idx0] - image_transformed
|
|
|
+ fraction_of_non_zero_pixels[1] = np.count_nonzero(image_transformed[:,:]<0.001)/262144
|
|
|
+
|
|
|
+ #plt.imshow(image_transformed)
|
|
|
+ #plt.show()
|
|
|
+ image_difference = image_difference[boarder_cut:-boarder_cut, boarder_cut:-boarder_cut]
|
|
|
+ image_difference = np.square(image_difference)
|
|
|
+ rmsd_rigid = np.sqrt(image_difference.sum()/(512 - 2 * boarder_cut)**2)
|
|
|
+ if (silent_mode!=True):
|
|
|
+ print("Fraction of non-zero pixels in 1 (mean rigid): ", fraction_of_non_zero_pixels[1], "Score", rmsd_rigid)
|
|
|
+
|
|
|
+ #plt.imshow(image_difference)
|
|
|
+
|
|
|
+
|
|
|
+### MEAN_ENH RIGID and AFFINE
|
|
|
+
|
|
|
+ reference_image = images_mean_enh[:, :, idx0]
|
|
|
+ initial_image = images_mean_enh[:, :, idx1]
|
|
|
+
|
|
|
+ # sx = ndimage.sobel(reference_image, axis=0, mode='constant')
|
|
|
+ # sy = ndimage.sobel(reference_image, axis=1, mode='constant')
|
|
|
+ # reference_image = np.hypot(sx, sy)
|
|
|
+
|
|
|
+ # sx = ndimage.sobel(initial_image, axis=0, mode='constant')
|
|
|
+ # sy = ndimage.sobel(initial_image, axis=1, mode='constant')
|
|
|
+ # initial_image = np.hypot(sx, sy)
|
|
|
+
|
|
|
+ boarder_cut = 100
|
|
|
+ sr = StackReg(StackReg.AFFINE)
|
|
|
+ tmats_affine_enh[idx0, idx1, :, :] = sr.register(reference_image, initial_image)
|
|
|
+
|
|
|
+ image_transformed = sr.transform(images_quality_check[:, :, idx1], tmats_affine_enh[idx0, idx1, :, :])
|
|
|
+ image_difference = images_quality_check[:, :, idx0] - image_transformed #TODO: delete image quality check! replace it with meanimage
|
|
|
+ fraction_of_non_zero_pixels[2] = np.count_nonzero(image_transformed[:,:]<0.001)/262144
|
|
|
+
|
|
|
+ #plt.imshow(image_transformed)
|
|
|
+ #plt.show()
|
|
|
+ image_difference = image_difference[boarder_cut:-boarder_cut, boarder_cut:-boarder_cut]
|
|
|
+ image_difference = np.square(image_difference)
|
|
|
+ rmsd_affine_enh = np.sqrt(image_difference.sum()/(512 - 2 * boarder_cut)**2)
|
|
|
+ if (silent_mode!=True):
|
|
|
+ print("Fraction of non-zero pixels in 2 (mean enh affine): ", fraction_of_non_zero_pixels[2],"Score:", rmsd_affine_enh)
|
|
|
+
|
|
|
+ sr = StackReg(StackReg.RIGID_BODY)
|
|
|
+ tmats_rigid_enh[idx0, idx1, :, :] = sr.register(reference_image, initial_image)
|
|
|
+ image_transformed = sr.transform(images_quality_check[:, :, idx1], tmats_rigid_enh[idx0, idx1, :, :])
|
|
|
+ image_difference = images_quality_check[:, :, idx0] - image_transformed
|
|
|
+ fraction_of_non_zero_pixels[0] = np.count_nonzero(image_transformed[:,:]<0.001)/262144
|
|
|
+
|
|
|
+ #plt.imshow(image_transformed)
|
|
|
+ #plt.show()
|
|
|
+ image_difference = image_difference[boarder_cut:-boarder_cut, boarder_cut:-boarder_cut]
|
|
|
+ image_difference = np.square(image_difference)
|
|
|
+ rmsd_rigid_enh = np.sqrt(image_difference.sum()/(512 - 2 * boarder_cut)**2)
|
|
|
+ if (silent_mode!=True):
|
|
|
+ print("Fraction of non-zero pixels in 0 (mean enh rigid): ", fraction_of_non_zero_pixels[0],"Score", rmsd_rigid_enh)
|
|
|
+
|
|
|
+ rmsds=[rmsd_rigid_enh,rmsd_rigid,rmsd_affine_enh,rmsd_affine]
|
|
|
+ tmatss=[tmats_rigid_enh[idx0, idx1, :, :],tmats_rigid[idx0, idx1, :, :],tmats_affine_enh[idx0, idx1, :, :],tmats_affine[idx0, idx1, :, :]]
|
|
|
+ methods=["rigid_mean_enh", "rigid_mean" ,"affine_mean_enh","affine_mean"]
|
|
|
+ #print(tmats_rigid_enh,tmats_rigid,tmats_affine_enh,tmats_affine)
|
|
|
+ #print(" ")
|
|
|
+ #best_method_idx = rmsds.index(min(rmsds))
|
|
|
+ #smaller_fraction_idx = fraction_of_non_zero_pixels.index(min(fraction_of_non_zero_pixels))
|
|
|
+ #smaller_fraction_idx = 1
|
|
|
+ #print(best_method_idx)
|
|
|
+ #print(smaller_fraction_idx)
|
|
|
+
|
|
|
+ list_of_methods=np.argsort(rmsds)
|
|
|
+
|
|
|
+ best_score[idx1, idx0] = np.sort(rmsds)[0]
|
|
|
+ best_score[idx0, idx1] = np.sort(rmsds)[0]
|
|
|
+
|
|
|
+ the_best_idx = list_of_methods[0]
|
|
|
+
|
|
|
+
|
|
|
+ if (fraction_of_non_zero_pixels[list_of_methods[0]] > 0.1):
|
|
|
+ print("Warning: alignment with the best method failed. The second best method is applied")
|
|
|
+ the_best_idx = list_of_methods[1]
|
|
|
+ if (fraction_of_non_zero_pixels[list_of_methods[1]] > 0.1):
|
|
|
+ print("Warning: alignment with the second best method failed. The 3rd best method is applied")
|
|
|
+ the_best_idx = list_of_methods[2]
|
|
|
+
|
|
|
+ best_method = methods[the_best_idx]
|
|
|
+ best_tmats[idx0, idx1, :, :]=tmatss[the_best_idx]
|
|
|
+ best_methods[idx1, idx0]=the_best_idx
|
|
|
+ best_methods[idx0, idx1]=the_best_idx
|
|
|
+
|
|
|
+
|
|
|
+ best_tmats[idx1, idx0, :, :]=np.linalg.inv(best_tmats[idx0, idx1, :, :])
|
|
|
+
|
|
|
+
|
|
|
+ if(idx0==idx1):
|
|
|
+ best_method="-,-"
|
|
|
+
|
|
|
+ if (silent_mode!=True):
|
|
|
+ print("{0:d}, {1:d}, {2:4.4f}, {3:4.4f}, {4:4.4f}, {5:4.4f}, {6:s}".format(idx0, idx1, rmsd_rigid_enh, rmsd_rigid, rmsd_affine_enh, rmsd_affine, best_method))
|
|
|
+
|
|
|
+ print("{0:d}, {1:d}, {2:4.4f}, {3:4.4f}, {4:4.4f}, {5:4.4f}, {6:s}".format(idx0, idx1, rmsd_rigid_enh, rmsd_rigid,
|
|
|
+ rmsd_affine_enh, rmsd_affine, best_method), file=f)
|
|
|
+
|
|
|
+ #print(" " + str(idx0) + "-" + str(idx1) + " " + str(rmsd_rigid_enh) + " " + str(rmsd_rigid) + " " + str(rmsd_affine_enh) + " " + str(rmsd_affine))
|
|
|
+ # plt.imshow(image_difference)
|
|
|
+
|
|
|
+ #plt.savefig(save_plots_path + "StackRegVisualInspection/" + file_title + "_d_reference_m_corrected.png")
|
|
|
+
|
|
|
+ #print(str(idx0) + '-' + str(idx1))
|
|
|
+ # save all the transformation matrices
|
|
|
+ if not os.path.exists(path4results+'StackReg'):
|
|
|
+ os.makedirs(path4results+'StackReg')
|
|
|
+ #print(best_tmats)
|
|
|
+ np.save(path4results+'StackReg/' + str(animal) + "_best_tmats", best_tmats)
|
|
|
+ np.save(path4results+'StackReg/' + str(animal) + "_best_methods", best_methods)
|
|
|
+ np.save(path4results+'StackReg/' + str(animal) + "_best_score", best_score)
|
|
|
+
|
|
|
+# if not os.path.exists('Q:/Personal/Mattia/Calcium Imaging/results/StackRegEnhImage'):
|
|
|
+# os.makedirs('Q:/Personal/Mattia/Calcium Imaging/results/StackRegEnhImage')
|
|
|
+# np.save('Q:/Personal/Mattia/Calcium Imaging/results/StackRegEnhImage/' + str(animal), tmats)
|
|
|
+# if not os.path.exists(save_plots_path+ 'StackRegAffine'):
|
|
|
+# os.makedirs(save_plots_path + 'StackRegAffine')
|
|
|
+# np.save(save_plots_path+ 'StackRegAffine/' + str(animal), tmats)
|
|
|
+f.close()
|
|
|
+
|
|
|
+# ### Install package for image comparison (similarity index)
|
|
|
+
|
|
|
+# In[79]:
|
|
|
+
|
|
|
+
|
|
|
+#get_ipython().system('conda install -c conda-forge imagehash --yes')
|
|
|
+#!pip install ImageHash # as alternative if anaconda is not installed
|
|
|
+
|
|
|
+
|
|
|
+# In[33]:
|
|
|
+
|
|
|
+from PIL import Image
|
|
|
+import imagehash
|
|
|
+
|
|
|
+
|
|
|
+for animal in animals:
|
|
|
+ print("Animal #", str(animal))
|
|
|
+
|
|
|
+ meta_animal = meta_data[meta_data['Mouse'] == animal]
|
|
|
+ recordings = meta_animal['Number']
|
|
|
+ index_similarity = np.zeros((np.shape(recordings)[0], np.shape(recordings)[0]))
|
|
|
+
|
|
|
+ cut = 100
|
|
|
+
|
|
|
+ images_mean = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+ images_mean_enh = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+
|
|
|
+ # load all (enhanced) images
|
|
|
+ for idx, recording in enumerate(recordings):
|
|
|
+ options = np.load(str(meta_data['Folder'][recording]) +
|
|
|
+ str(meta_data['Subfolder'][recording]) +
|
|
|
+ #str(int(meta_data['Recording idx'][recording])) +
|
|
|
+ '/suite2p/plane0/ops.npy',
|
|
|
+ allow_pickle=True)
|
|
|
+ # mean image or mean enhanced image
|
|
|
+ images_mean[:, :, idx] = options.item(0)['meanImg']
|
|
|
+ images_mean_enh[:, :, idx] = options.item(0)['meanImgE']
|
|
|
+
|
|
|
+
|
|
|
+ for idx0 in range(np.shape(images_mean)[2]):
|
|
|
+ for idx1 in range(idx0, np.shape(images_mean)[2]):
|
|
|
+ hash1=imagehash.average_hash(Image.fromarray(images_mean[cut:-cut, cut:-cut, idx0]))
|
|
|
+ otherhash=imagehash.average_hash(Image.fromarray(images_mean[cut:-cut, cut:-cut, idx1]))
|
|
|
+ index_similarity[idx0,idx1] = (hash1 - otherhash)
|
|
|
+ index_similarity[idx1,idx0] = (hash1 - otherhash)
|
|
|
+ #index_similarity = (np.max(index_similarity)-index_similarity)/np.max(index_similarity)
|
|
|
+
|
|
|
+ best_tmats = np.load(path4results+'StackReg/' + str(animal) + "_best_tmats.npy")
|
|
|
+ best_score = np.load(path4results+'StackReg/' + str(animal) + "_best_score.npy")
|
|
|
+
|
|
|
+ metric_best_tmats = np.abs(best_tmats)
|
|
|
+ metric_best_tmats = metric_best_tmats.sum(axis=(2,3))
|
|
|
+ metric_best_tmats = np.max(metric_best_tmats) - metric_best_tmats
|
|
|
+
|
|
|
+ metric_best_score = (np.max(best_score)-best_score)/np.max(best_score)
|
|
|
+ #plt.xticks(np.arange(0, np.shape(images_mean)[2], 1));
|
|
|
+ #plt.yticks(np.arange(0, np.shape(images_mean)[2], 1));
|
|
|
+
|
|
|
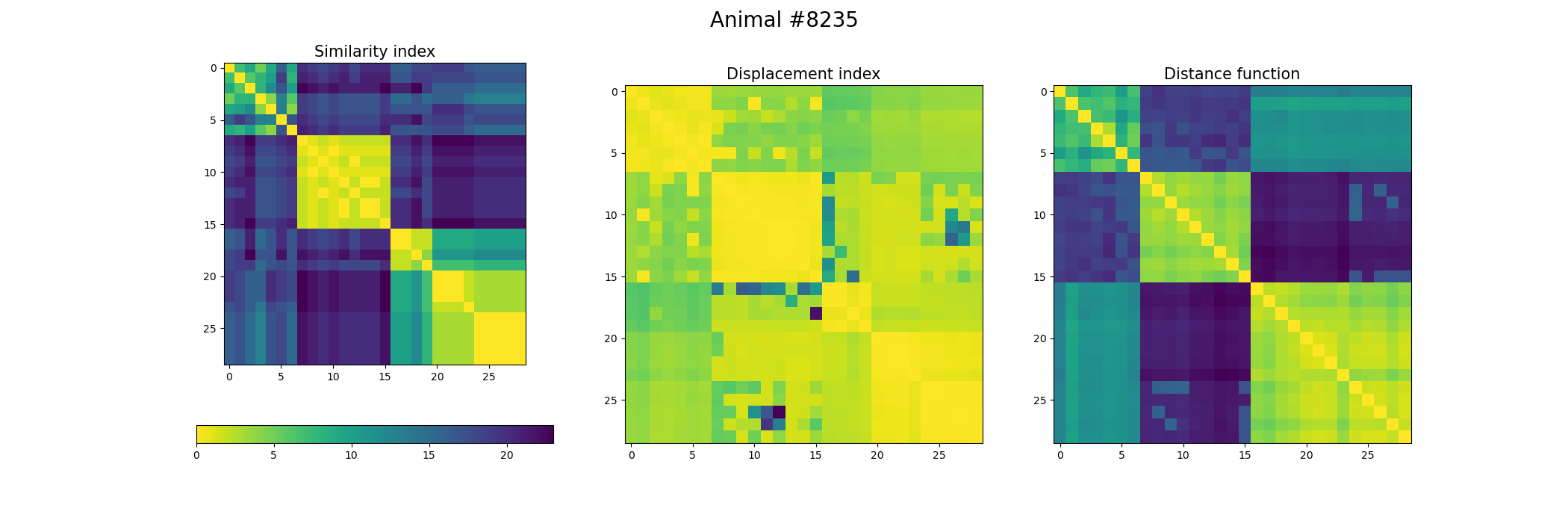
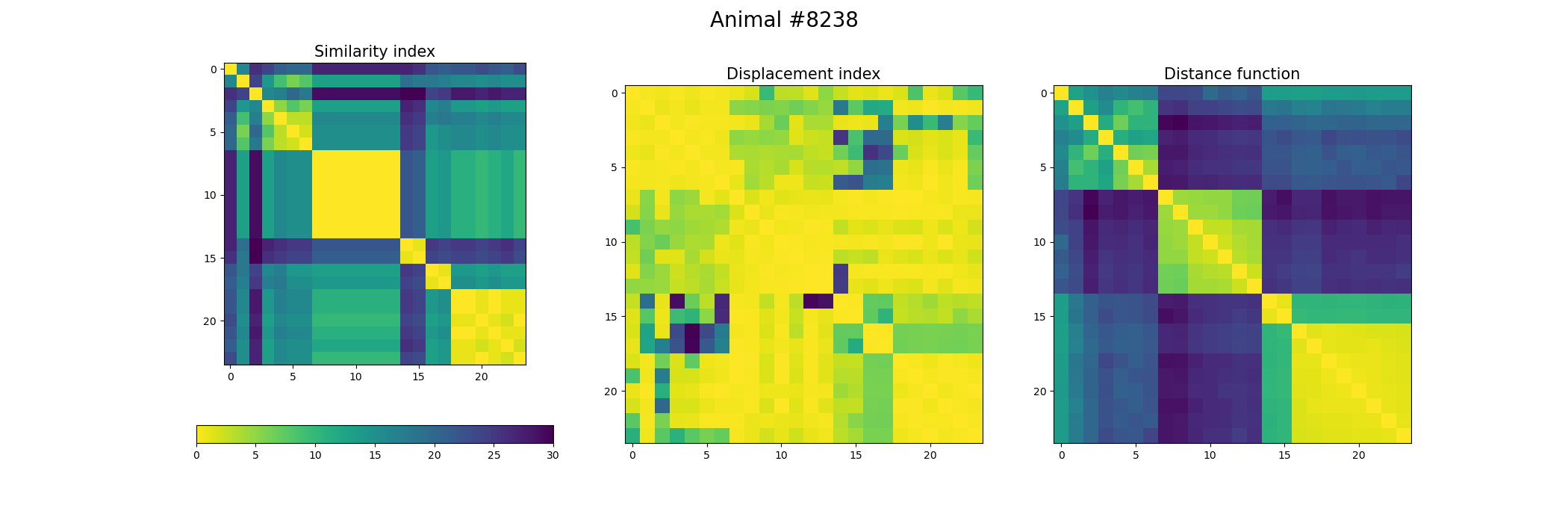
+ fig, ax = plt.subplots(1,3, figsize=(21, 7))
|
|
|
+
|
|
|
+ index_similarity[index_similarity > 30] = 30
|
|
|
+ image = ax[0].imshow(index_similarity,cmap='viridis_r') #,cmap=cmap, norm=norm
|
|
|
+ cbar = fig.colorbar(image, ax=ax[0], orientation='horizontal', fraction=.1)
|
|
|
+
|
|
|
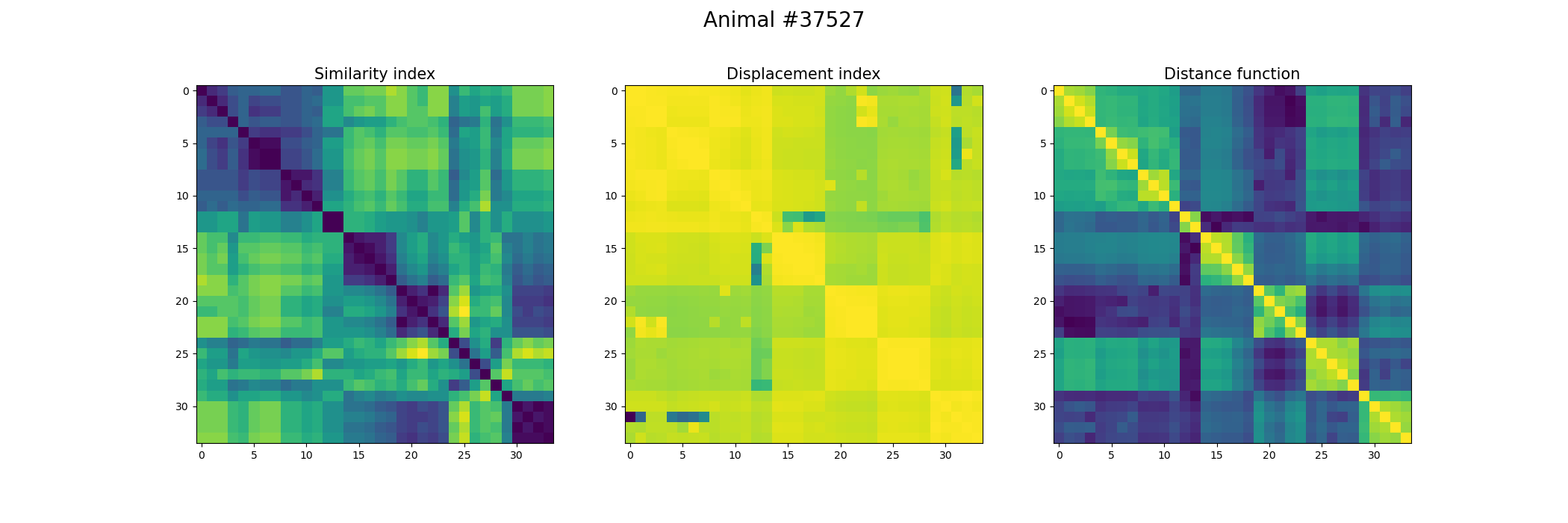
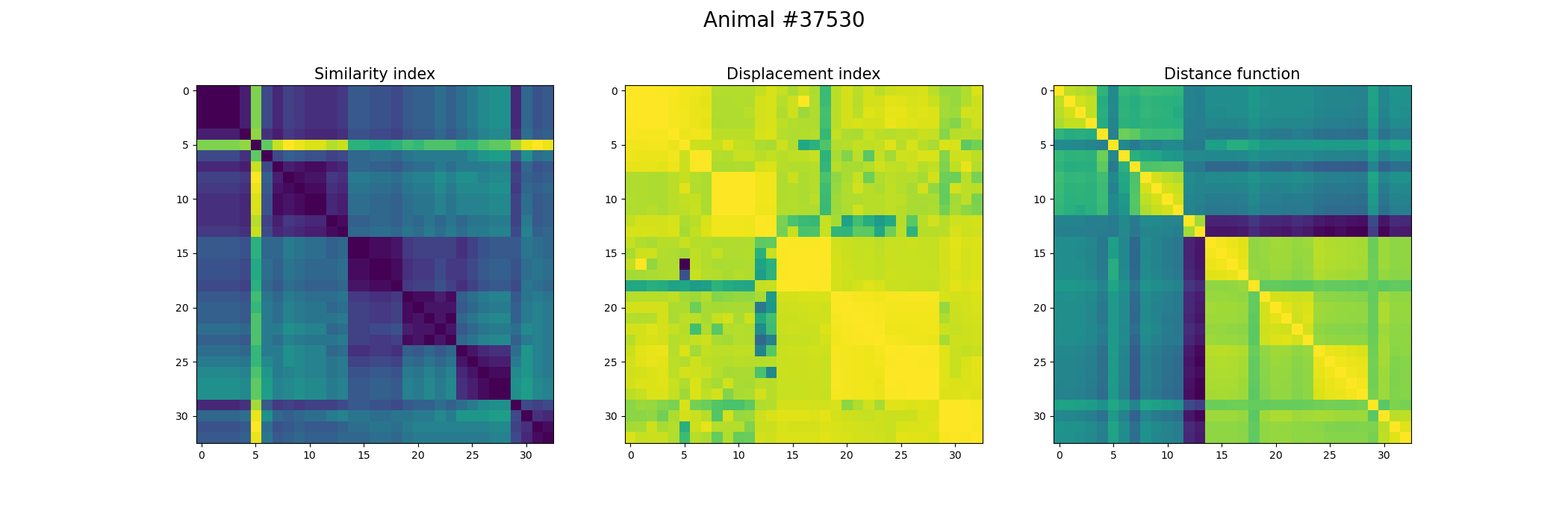
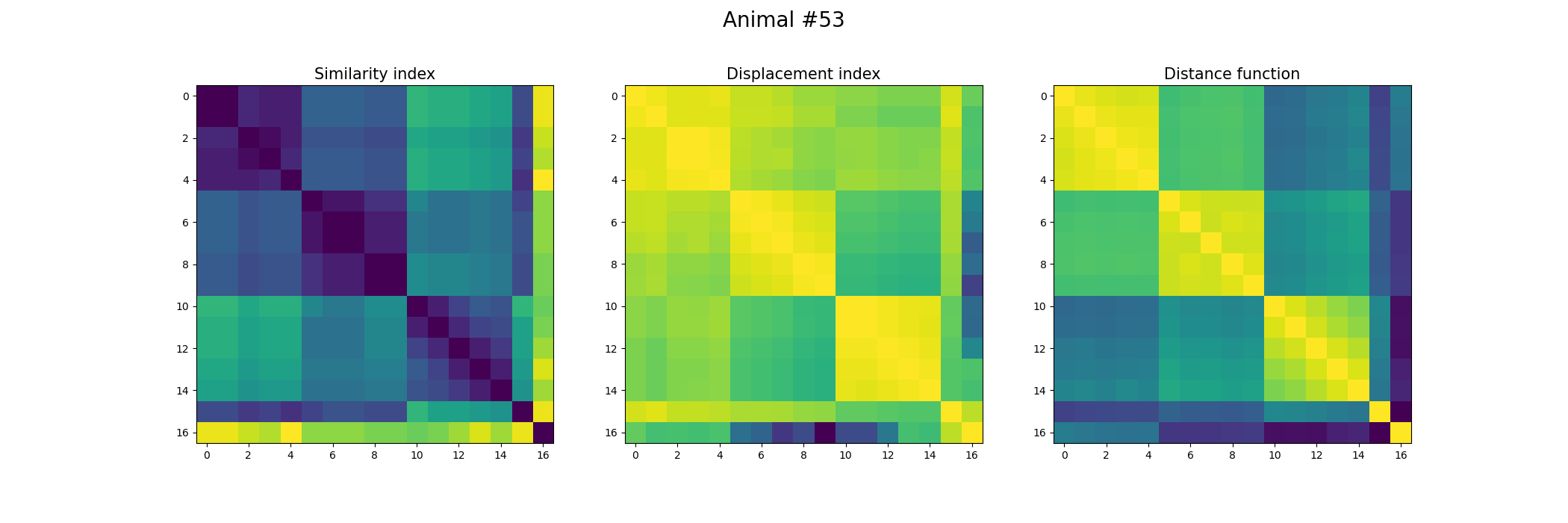
+ ax[0].set_title("Similarity index", fontsize = 15)
|
|
|
+ ax[1].imshow(metric_best_tmats)
|
|
|
+ ax[1].set_title("Displacement index", fontsize = 15)
|
|
|
+ ax[2].imshow(metric_best_score)
|
|
|
+ ax[2].set_title("Distance function", fontsize = 15)
|
|
|
+ fig.suptitle('Animal #' + str(animal), fontsize = 20)
|
|
|
+
|
|
|
+ plt.savefig("./FOV_Validation_" + str(animal) + ".png")
|
|
|
+
|
|
|
+
|
|
|
+# ### Plot all corrected FOV's for comparison (optional)
|
|
|
+#
|
|
|
+# **The running takes considerable amount of time!**
|
|
|
+
|
|
|
+# In[23]:
|
|
|
+
|
|
|
+
|
|
|
+for animal in animals:
|
|
|
+
|
|
|
+ tmats_loaded = np.load(path4results + 'StackReg/' + str(animal) + "_best_tmats" + '.npy')
|
|
|
+ meta_animal = meta_data[meta_data['Mouse'] == animal]
|
|
|
+ recordings = meta_animal['Number']
|
|
|
+ images = np.zeros((512, 512, np.shape(meta_animal)[0]))
|
|
|
+
|
|
|
+ images_mean = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+ images_mean_enh = np.zeros((512, 512, np.shape(recordings)[0]))
|
|
|
+
|
|
|
+ # load all (enhanced) images
|
|
|
+ for idx, recording in enumerate(recordings):
|
|
|
+ options = np.load(meta_data['Folder'][recording] +
|
|
|
+ meta_data['Subfolder'][recording] +
|
|
|
+ str(int(meta_data['Recording idx'][recording])) +
|
|
|
+ '/suite2p/plane0/ops.npy',
|
|
|
+ allow_pickle=True)
|
|
|
+ # mean image or mean enhanced image
|
|
|
+ images_mean[:, :, idx] = options.item(0)['meanImg']
|
|
|
+ images_mean_enh[:, :, idx] = options.item(0)['meanImgE']
|
|
|
+ #cut_boarders=50
|
|
|
+ #quality check
|
|
|
+ #images_quality_check[:, :, idx] = options.item(0)['meanImg']
|
|
|
+
|
|
|
+
|
|
|
+ # loop through every pair and compute the transformation matrix
|
|
|
+
|
|
|
+ conditions = [meta_data['Condition'][recording] for recording in recordings]
|
|
|
+ recording_idx = [meta_data['Recording idx'][recording] for recording in recordings]
|
|
|
+
|
|
|
+ for idx0 in range(np.shape(images_mean)[2]):
|
|
|
+ #if (idx0!=14):
|
|
|
+ #continue
|
|
|
+ for idx1 in range(idx0, np.shape(images_mean)[2]):
|
|
|
+ #if (idx1!=16):
|
|
|
+ #continue
|
|
|
+
|
|
|
+ reference_image = images_mean_enh[:, :, idx0]
|
|
|
+ initial_image = images_mean_enh[:, :, idx1]
|
|
|
+
|
|
|
+ if not os.path.exists(save_plots_path + 'StackRegVisualInspection/'):
|
|
|
+ os.makedirs(save_plots_path + 'StackRegVisualInspection/')
|
|
|
+ if not os.path.exists(save_plots_path + 'StackRegVisualInspection/' + str(animal) + '/'):
|
|
|
+ os.makedirs(save_plots_path + 'StackRegVisualInspection/' + str(animal) + '/')
|
|
|
+
|
|
|
+
|
|
|
+ plt.imshow(reference_image)
|
|
|
+
|
|
|
+ # image_title = meta_data['Subfolder'][recording][:-1] + str(meta_data['Recording idx'][recording]) + "\n" + "_condition_" + \
|
|
|
+ # meta_data['Condition'][recording]
|
|
|
+ # plt.title(image_title)
|
|
|
+ #file_title = meta_data['Subfolder'][recording][:-1] + str(
|
|
|
+ # meta_data['Recording idx'][recording]) + "_condition_" + \
|
|
|
+ # meta_data['Condition'][recording] + "_" + str(idx0) + "_" + str(idx1)
|
|
|
+
|
|
|
+ file_title = str(str(idx0) + '_' + str(idx1) + '_' + conditions[idx0]) + '_' + str(recording_idx[idx0]) + '_' + str(conditions[idx1]) + "_" + str(recording_idx[idx1])
|
|
|
+ print(file_title)
|
|
|
+
|
|
|
+ plt.savefig(save_plots_path + "StackRegVisualInspection/" + str(animal) + '/' + file_title + "_b_reference.png")
|
|
|
+ #sx = ndimage.sobel(images[:, :, idx1], axis=0, mode='constant')
|
|
|
+ #sy = ndimage.sobel(images[:, :, idx1], axis=1, mode='constant')
|
|
|
+ #sob = np.hypot(sx, sy)
|
|
|
+ #plt.imshow(sob)
|
|
|
+ plt.imshow(initial_image)
|
|
|
+ plt.savefig(save_plots_path + "StackRegVisualInspection/" + str(animal) + '/' + file_title + "_a_initial.png")
|
|
|
+ #grad = np.gradient(images[:, :, idx1])
|
|
|
+
|
|
|
+ #print(images[50:55, 50:55, idx1].shape)
|
|
|
+ #grad = np.gradient(images[50:55, 50:55, idx1])
|
|
|
+ #print(images[50:55, 50:55, idx1])
|
|
|
+ #print(" ")
|
|
|
+ #print(grad)
|
|
|
+
|
|
|
+ #image_inversed = sr.transform(reference_image, best_tmats[idx1, idx0, :, :])
|
|
|
+ #plt.imshow(image_inversed)
|
|
|
+ #plt.savefig(save_plots_path + "StackRegVisualInspection/" + str(animal) + '/' + file_title + "_1_inversed.png")
|
|
|
+
|
|
|
+ #sx = ndimage.sobel(images[:, :, idx1], axis=0, mode='constant')
|
|
|
+ #sy = ndimage.sobel(images[:, :, idx1], axis=1, mode='constant')
|
|
|
+ #sob = np.hypot(sx, sy)
|
|
|
+ #plt.imshow(images[:, :, idx1])
|
|
|
+ #plt.savefig(save_plots_path + "StackRegVisualInspection/" + file_title + "_sobel.png")
|
|
|
+ image_corrected = sr.transform(initial_image, tmats_loaded[idx0, idx1, :, :])
|
|
|
+ plt.imshow(image_corrected)
|
|
|
+ plt.savefig(save_plots_path + "StackRegVisualInspection/" + str(animal) + '/' + file_title + "_c_corrected.png")
|
|
|
+ #image_difference = images_quality_check[:, :, idx0] - sr.transform(images_quality_check[:, :, idx1], best_tmats[idx0, idx1, :, :])
|
|
|
+ #image_difference = reference_image - image_corrected
|
|
|
+ #plt.imshow(image_difference)
|
|
|
+ #plt.savefig(save_plots_path + "StackRegVisualInspection/" + str(animal) + '/' + file_title + "_d_reference_m_corrected.png")
|
|
|
+
|
|
|
+
|
|
|
+# In[9]:
|
|
|
+
|
|
|
+
|
|
|
+###TODO Seaborn Scatterplot heatmap could be a nice option for plotting
|
|
|
+
|
|
|
+
|
|
|
+# In[ ]:
|
|
|
+
|
|
|
+
|
|
|
+
|