|
|
@@ -0,0 +1,603 @@
|
|
|
+\RequirePackage{fix-cm}
|
|
|
+%\documentclass{article}
|
|
|
+%\documentclass{svjour3} % onecolumn (standard format)
|
|
|
+%\documentclass[smallcondensed]{svjour3} % onecolumn (ditto)
|
|
|
+\documentclass[smallextended]{svjour3} % onecolumn (second format)
|
|
|
+%\documentclass[twocolumn]{svjour3} % twocolumn
|
|
|
+\usepackage[utf8]{inputenc}
|
|
|
+
|
|
|
+
|
|
|
+\usepackage[margin=1in]{geometry}
|
|
|
+\usepackage[toc]{appendix}
|
|
|
+\usepackage{natbib}
|
|
|
+
|
|
|
+\usepackage{booktabs}
|
|
|
+\usepackage{hyperref}
|
|
|
+
|
|
|
+
|
|
|
+\makeatletter
|
|
|
+\newcommand\footnoteref[1]{\protected@xdef\@thefnmark{\ref{#1}}\@footnotemark}
|
|
|
+\makeatother
|
|
|
+
|
|
|
+
|
|
|
+\usepackage{tikz}
|
|
|
+\usetikzlibrary{arrows.meta,positioning,calc,shapes}
|
|
|
+\usepackage[edges]{forest}
|
|
|
+\definecolor{foldercolor}{RGB}{124,166,198}
|
|
|
+\newcommand{\inputTikZ}[2]{%
|
|
|
+ \scalebox{#1}{\input{#2}}
|
|
|
+}
|
|
|
+\usepackage{subfig}
|
|
|
+\usepackage[outdir=./plots]{epstopdf}
|
|
|
+\usepackage{textcomp}
|
|
|
+
|
|
|
+
|
|
|
+\usepackage[Symbol]{upgreek}
|
|
|
+
|
|
|
+\graphicspath{.}
|
|
|
+
|
|
|
+\title{Managing, storing and sharing long-form recordings and their annotations}
|
|
|
+
|
|
|
+\author{%
|
|
|
+Lucas Gautheron \and Nicolas Rochat \and Alejandrina Cristia
|
|
|
+}
|
|
|
+
|
|
|
+\institute{
|
|
|
+Laboratoire de Sciences Cognitives et de Psycholinguistique, Département d'Etudes cognitives, ENS, EHESS, CNRS, PSL University, Paris, France.
|
|
|
+\email{lucas.gautheron@gmail.com}
|
|
|
+}
|
|
|
+
|
|
|
+\journalname{Language Resources and Evaluation}
|
|
|
+
|
|
|
+\date{}
|
|
|
+
|
|
|
+\begin{document}
|
|
|
+
|
|
|
+\maketitle
|
|
|
+
|
|
|
+\abstract{
|
|
|
+The technique of \textit{in situ}, long-form recordings is gaining momentum in different fields of research, notably linguistics and pathology. This method, however, poses several technical challenges, some of which are amplified by the peculiarities of the data, including their sensitiveness and their volume. In the following paper, we begin by outlining the problems related to the management, storage, and sharing of the corpora produced using this technique. We then go on to propose a multi-component solution to these problems, in the specific case of daylong recordings of children. As part of this solution, we release \emph{ChildProject}, a python package to perform the operations typically required to work with such datasets. The package also provides built-in functions to evaluate the annotations using a number of measures commonly used in speech processing and linguistics. Our proposal, as we argue, could be generalized to broader populations.
|
|
|
+}
|
|
|
+
|
|
|
+\keywords{daylong recordings, speech data management, data distribution, annotation evaluation, inter-rater reliability}
|
|
|
+
|
|
|
+
|
|
|
+%\tableofcontents
|
|
|
+
|
|
|
+
|
|
|
+%\begin{itemize}
|
|
|
+% \item adding the large amounts of data into the problem space ? (because it means high storage costs, delivery difficulties etc.)
|
|
|
+% \item More emphasis on reproducibility and how DataLad is helpful with that
|
|
|
+% \item Find some rationale to decide when to refer to git-annex or when to refer to DataLad, repository vs dataset etc.
|
|
|
+% \item Referring to git files (as opposed to git annex files) as "text files" is ambiguous as annotations are text files, but usually stored in the annex. But "git files" is ambiguous to as it could mean everything inside of .git...
|
|
|
+% \item OSF integration: example 3 ?
|
|
|
+% \item Language Archive
|
|
|
+% \item FAIR
|
|
|
+% \item coin \citep{Gorgolewski2016}
|
|
|
+%\end{itemize}
|
|
|
+
|
|
|
+\section{Introduction}
|
|
|
+
|
|
|
+Long-form recordings are those collected over extended periods of time, typically via a wearable. Although the technique was used with normotypical adults decades ago \citep{ear1,ear2}, it became widespread in the study of early childhood over the last 15 years or so. The LENA Foundation created a hardware-software combination that illuminated the potential of this technique for theoretical and applied purposes (e.g., \citealt{christakis2009audible,warlaumont2014social}). More recently, such data is being discussed in the context of neurological disorders (e.g., \citealt{riad2020vocal}). In this article, we define the unique space of difficulties surrounding long-form recordings, and introduce a python package that provides practical solutions, with a focus on child-centered recordings. We end by discussing ways in which these solutions could be generalized to other populations.
|
|
|
+
|
|
|
+\section{Problem space}\label{section:problemspace}
|
|
|
+
|
|
|
+Management of scientific data is a long-standing issue, which has been subject to substantial progress in the recent years. For instance, FAIR principles \citep{Wilkinson2016} - where the initials stand for Findability, Accessibility, Interoperability, and Reusability - have been proposed to help increase the usefulness of data and data analysis pipelines. Similarly, databases implementing these practices have emerged, such as Dataverse \citep{dataverse} and Zenodo \citep{zenodo}. The method of daylong recordings should incorporate such methodological advances. It should be noted, however, that some of the difficulties surrounding the management of corpora of daylong recordings are more idiosyncratic to this technique, and therefore require specific solutions to be developed. Below, we list some of the challenges researchers engaging in the technique of long-form recordings in naturalistic environments are likely to face.
|
|
|
+
|
|
|
+\subsubsection*{The need for standards}
|
|
|
+
|
|
|
+Extant datasets rely on a wide variety of metadata structures, file formats, and naming conventions. For instance, some data from long-form recordings have been archived publicly on Databrary (such as the ACLEW starter set \citep{starter}) and HomeBank (including the VanDam Daylong corpus from \citealt{vandam-day}). Table \ref{table:datasets} shows some divergence across the two. As a result of this divergence, each lab finds itself re-inventing the wheel. For instance, the HomeBankCode organization \footnote{\url{https://github.com/homebankcode/}} contains at least 4 packages that do more or less the same operations of e.g. aggregating how much speech was produced in each recording, implemented in different languages (MatLab, R, perl, and Python). This divergence may also hide different operationalizations, rendering comparisons across labs fraught, effectively diminishing replicability.\footnote{\textit{Replicability} is typically defined as the effort to re-do a study on a new sample, whereas \textit{reproducibility} relates to re-doing the exact same analyses in the exact same data. Reproducibility is addressed in another section.}
|
|
|
+
|
|
|
+Designing pipelines and analyses that are consistent across datasets requires standards in how the datasets are structured. Although this may represent an initial investment, such standards facilitate the pooling of research efforts, by allowing labs to benefit from code developed in other labs. Additionally, this field operates increasingly via collaborative cross-lab efforts. For instance, the ACLEW project\footnote{\url{sites.google.com/site/aclewdid}} involved 9 principal investigators (PIs) from 5 different countries, who needed a substantive initial investment to agree on a standard organization for their 6 corpora. We expect even larger collaborations to emerge in the future, a move that would be benefited by standardization, as exemplified by the community that emerged around CHILDES for short-form recordings \citep{macwhinney2000childes}.
|
|
|
+
|
|
|
+\begin{table}
|
|
|
+\centering
|
|
|
+\begin{tabular}{@{}lll@{}}
|
|
|
+\toprule
|
|
|
+ & ACLEW starter & Van Dam \\ \midrule
|
|
|
+\begin{tabular}[t]{@{}l@{}}Audio's scope\end{tabular} & 5-minute clips & Full day \\
|
|
|
+\begin{tabular}[t]{@{}l@{}}Automated annotations'\\format\end{tabular} & none & LENA \\
|
|
|
+\begin{tabular}[t]{@{}l@{}}Automated annotations'\\format\end{tabular} & .eaf & .cha \\
|
|
|
+Annotations' scope & only clips & Full day \\
|
|
|
+Metadata & none & excel \\ \bottomrule
|
|
|
+\end{tabular}
|
|
|
+\caption{\textbf{Divergences between the \cite{starter} and \cite{vandam-day} datasets}. Audios' scope indicates the size of the audio that has been archived: all recordings last for a full day, but for ACLEW starter, three five-minute clips were selected from each child. Automated annotations' format indicates what software was used to annotate the audio automatically. Annotations' scope shows the scope of human annotation. Metadata indicates whether information about the children and recording were shared, and in what format.}
|
|
|
+\label{table:datasets}
|
|
|
+\end{table}
|
|
|
+
|
|
|
+
|
|
|
+\subsubsection*{Keeping up with updates and contributions}
|
|
|
+
|
|
|
+Datasets are not frozen. Rather, they are continuously enriched through annotations provided by humans or new algorithms. Human annotations may also undergo corrections, as errors are discovered. The process of collecting the recordings may also require a certain amount of time, as they are progressively returned by the field workers or the participants themselves. In the case of longitudinal studies, supplementary audio data may accumulate over several years. Researchers should be able to keep track of these changes while also upgrading their analyses. Moreover, several collaborators may be brought to contribute work to the same dataset simultaneously.
|
|
|
+
|
|
|
+To take the example of ACLEW, PIs first annotated in-house a random selection of 2-minute clips for 10 children. They then exchanged some of these audios so that the annotators in another lab re-annotated the same data, for the purposes of inter-rater reliability. This revealed divergences in definitions, and all datasets needed to be revised. Finally, a second sample of 2-minute clips with high levels of speech activity were annotated -- and another process of reliability was performed.
|
|
|
+
|
|
|
+\subsubsection*{Delivering large amounts of data}
|
|
|
+
|
|
|
+Considering typical values for the bit depth and sampling rates of the recordings -- 16 bits and 16 kilohertz respectively -- yield a throughput of approximately three gigabytes per day of audio. Although there is a great deal of variation, past studies often involved at least 30 recording days (e.g., 3 days for each of 10 children). The trend, however, is for datasets to be larger; for instance, last year, we collected 2 recordings from about 200 children. Such datasets may exceed one terabyte. Moreover, these recordings can be associated with annotations spread across thousands of files. In the ACLEW example just discussed, there was one .eaf file per human annotator per type of annotation (i.e., random, high speech, random reliability, high speech reliability). In addition, the full day was analyzed with between 1 and 4 automated routines. Thus, for each recording day there were 8 annotation files, leading to 5 corpora $\times$ 10 children $\times$ 8 annotation = 400 annotation files. Other researchers will use one annotation file per clip selected for annotation, which quickly adds up to thousands of files. Even a small processing latency may add up to significant overheads while gathering so many files.
|
|
|
+Data-access should be doable programmatically, and users should be able to download only the data that they need for their analysis.
|
|
|
+
|
|
|
+
|
|
|
+\subsubsection*{Privacy}
|
|
|
+
|
|
|
+Long-form recordings are sensitive; they contain identifying and personal information about the participating family. In some cases, for instance if the family goes shopping and forgets to notify those around them, recordings could capture conversations which involve people who are unaware that they are being recorded. In addition, they may be subject to regulations, such as the European GDPR, the American HIPAA, and, depending on the place of collection and/or storage, laws on biometric data.
|
|
|
+
|
|
|
+However, although the long-form recordings are sensitive, many of the data types derived from them are not. With appropriate file-naming and meta-data practices, it is effectively possible to completely deidentify automated annotations (which at present never include automatic speech recognition). It is also often possible to deidentify human annotations, except when these involve transcribing what participants said, since participants will use personal names and reveal other personal details. Nonetheless, since this particular case involves a human doing the annotation, this human can be trained to modify the record (e.g., replace personal names with foils) and/or tag the annotation as sensitive and not to be openly shared (a practice called vetting \citep{Cychosz2020}.)
|
|
|
+
|
|
|
+Therefore, the ideal storing-and-sharing strategy should naturally enforce security and privacy safeguards by implementing access restrictions adapted to the level of confidentiality of the data.
|
|
|
+
|
|
|
+\subsubsection*{Long-term availability}
|
|
|
+
|
|
|
+The collection of long-form recordings requires a considerable level of investment to explain the technique to families and communities, ensure a secure data management system, and, in the case of remote populations, access to and from the site. In our experience, one data collection trip to a field site costs about 15 thousand US\$.\footnote{This grossly underestimates overall costs, because the best way to do any kind of field research is by maintaining strong bonds with the community and helping them in other ways throughout the year, rather than only during our visits. A successful example for this is that of the UNM-UCSB Tsimane' Project (\url{http://tsimane.anth.ucsb.edu/}), which has been collaborating with the Tsimane' population since 2001. They are currently funded by a 5-year, 3-million US\$ NIH grant \url{https://reporter.nih.gov/project-details/9538306}}. These data are precious not only because of the investment that has gone into them, but because they capture slices of life at a given point in time, which is particularly informative in the case of populations that are experiencing market integration or other forms of societal change -- which today is most or all populations. Moreover, some communities who are collaborating in such research speak languages that are minority languages in the local context, and thus at a potential risk for being lost in the future. The conservation of naturalistic speech samples of children's language acquisition throughout a normal day could be precious to fuel future efforts of language revitalization \citep{Nee2021}. It would therefore be particularly damaging to lose such data prematurely, from a financial, a scientific, and a human standpoints.
|
|
|
+
|
|
|
+In addition, one advantage of daylong recordings over other observational methods such as parental reports is that they can be re-exploited at later times to observe behaviors that had not been foreseen at the time of data collection. This implies that their interest partly lies in long-term re-usability.
|
|
|
+
|
|
|
+Moreover, even state-of-the-art speech processing tools still perform poorly on daylong recordings, due to their intrinsic noisy nature \citep{casillas2019step}. As a result, taking full advantage of present data will necessitate new or improved computational models, which may take years to develop. For example, the DIHARD Challenge series has been running for three years in a row, and documents the difficulty of making headway with complex audio data \citep{ryant2018first,ryant2019second,ryant2020third}. For instance, the best submission for speaker diarization in their meeting subcorpus achieved about 35\% Diarization Error Rate in 2018 and 2019, with improvements seen only in 2020, when the best system scored 20\% Diarization Error Rate (Neville Ryant, personal communication, 2021-04-09). Other tasks are progressing much more slowly. For instance, the best performance in a classifier for deciding whether adult speech was addressed to the child or to an adult scored about 70\% correct in 2017 \citep{schuller2017interspeech} -- but nobody has been able to beat this record since. Recordings should therefore remain available for long periods of time -- potentially decades --, thus increasing the risk for data loss to occur at some point in their lifespan. For these reasons, the reliability of the storage design is critical, and redundancy is most certainly required. Likewise, persistent URLs may be needed in order to ensure the long-term accessibility of the datasets.
|
|
|
+
|
|
|
+\subsubsection*{Findability}
|
|
|
+
|
|
|
+FAIR Principles include findability and accessibility. A crucial aspect of findability of datasets involves their being indexed in ways that potential re-users can discover them. As we will mention below, there is one archiving option that is specific for long-form recordings, which thus makes any corpora hosted on there easily discoverable by other researchers working with that technique; and another specializing on child development, which can interest the developmental science community. However, the standard practice today is that data are archived in either one or another of these repositories, despite the fact that if an instance of the corpus were visible from one of these archives, the dataset would be overall more easily discovered. Additionally, we are uncertain the extent to which these highly re-usable long-form recordings are visible to researchers more broadly interested in spoken corpora and/or naturalistic human behavior and/or other topics that could be studied in such data. In fact, one can conceive of a future in which the technique begins to be used with people of different ages, in which case a system that allows users to discover other datasets based on relevant metadata would be ideal: For some research purposes (e.g., the study of source separation) any recording may be useful, whereas for others (neurodegenerative disorders, early language acquisition) only some ages would. In any case, options exist to allow accessibility once a dataset is archived in one of those archives.
|
|
|
+
|
|
|
+\subsubsection*{Reproducibility}
|
|
|
+
|
|
|
+Independent verification of results by a third party can be facilitated by improving the \emph{reproducibility} of the analyses, i.e. by providing third-parties with enough data and information to re-derive claimed results. This itself maybe be challenging for a number of reasons, including the variety of software requirements, unclear data dependencies, or insufficiently documented steps. Sharing data sets and analyses is more complex than delivering a collection of static files; all the information that is necessary in order to re-execute any intermediate step of the analysis should also be adequately conveyed.
|
|
|
+
|
|
|
+
|
|
|
+
|
|
|
+\subsubsection*{Current archiving options}
|
|
|
+
|
|
|
+The field of child-centered long-form recordings benefited from a purpose-built scientific archive from very early on. HomeBank \cite{vandam2016homebank} builds on the same architecture as CHILDES \cite{MacWhinney2000} and other TalkBank corpora. Although this architecture served the purposes of the language-oriented community well for short recordings, there are numerous issues when using it for long-form recordings. To begin with, curators do not directly control their datasets' contents and structures, and if a curator wants to make a modification, they need to ask the HomeBank management team to make it for them. Similarly, other collaborators who spot errors cannot correct them directly, but again must request changes be made by the HomeBank management team. Only one type of annotation is innately managed, and that is CHAT \cite{MacWhinney2000}, which is ideal for transcriptions of what was said. However, transcription is less central to studies of long-form audio.
|
|
|
+
|
|
|
+Other options have been used by researchers in the community, including OSF, Databrary, and the Language Archive. Detailing all their features is beyond the scope of the present paper, but some discussion can be found in \cite{casillas2019step}. For our purposes, the key issue to bear in mind is that none of these archives supports well the very large audio files found in long-form corpora. These limitations have brought us to envision a new strategy for sharing these datasets.
|
|
|
+
|
|
|
+ \subsubsection*{Our proposal}
|
|
|
+
|
|
|
+We propose a storing-and-sharing method designed to address the challenges outlined above simultaneously. It can be noted that these problems are, in many respects, similar to those faced by researchers in neuroimaging, a field which has long been confronting the need for reproducible analyses on large datasets of potentially sensitive data \citep{Poldrack2014}.
|
|
|
+Their experience may, therefore, provide precious insight for linguists, psychologists, and developmental scientists engaging with the big-data approach of daylong recordings.
|
|
|
+For instance, in the context of neuroimaging, \citet{Gorgolewski2016} have argued in favor of ``machine-readable metadata'', standard file structures and metadata, as well as consistency tests. Similarly, \citet{Eglen2017} have recommended the use of formatting standard, version control, and continuous testing. In the following, we will demonstrate how all of these practices have been implemented in our proposed design.
|
|
|
+
|
|
|
+Albeit designed for child-centered daylong recordings, we believe our solution could be replicated across a wider range of datasets with constraints similar to those exposed above. Furthermore, our approach is flexible and leaves room for customization.
|
|
|
+
|
|
|
+This solution relies on four main components, each of which is conceptually separable from the others: i) a standardized data format, optimized for child-centered long-form recordings; \citep{hanke_defense_2021}; ii) ChildProject, a python package to perform basic operations on these datasets; iii) DataLad, ``a decentralized system for integrated discovery, management, and publication of digital objects of science'' iv) GIN, a live archiving option for storage and distribution. Our choice for each one of these components can be revisited based on the needs of a project and/or as other options appear. Table \ref{table:components} summarises which of these components help address each of the challenges listed in Section \ref{section:problemspace}.
|
|
|
+
|
|
|
+\begin{table*}[ht]
|
|
|
+\centering
|
|
|
+\begin{tabular}{@{}l|llll@{}}
|
|
|
+\toprule
|
|
|
+\textbf{Problem} &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}\textbf{ChildProject}\\(Section \ref{section:childproject})\end{tabular} &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}\textbf{DataLad}\\(Section \ref{section:datalad})\end{tabular} &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}\textbf{GIN}\\(Section \ref{section:gin})\end{tabular} \\ \midrule
|
|
|
+The need for standards &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}documented standards;\\tests;\\conversion routines\end{tabular} &
|
|
|
+ &
|
|
|
+ \\ \midrule
|
|
|
+\begin{tabular}[t]{@{}l@{}}Keeping up with updates\\ and contributions\end{tabular} &
|
|
|
+ &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}version control\\(git)\end{tabular} &
|
|
|
+ git repository host
|
|
|
+ \\ \midrule
|
|
|
+\begin{tabular}[t]{@{}l@{}}Delivering large amounts\\ of data\end{tabular} &
|
|
|
+ parallelised processing &
|
|
|
+ git-annex &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}git-annex compatible;\\ high storage capacity;\\ parallelised operations\end{tabular}
|
|
|
+ \\ \midrule
|
|
|
+Ensuring privacy &
|
|
|
+ &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}private sub-datasets;\\ private remotes;\\
|
|
|
+ path-based\\or metadata-based\\
|
|
|
+ storage rules;\end{tabular} &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}Access Control Lists;\\SSH authentication\end{tabular}
|
|
|
+ \\ \midrule
|
|
|
+Long-term storage & \begin{tabular}[t]{@{}l@{}}tests\\(ensure integrity;\\
|
|
|
+detect missing files)\end{tabular}
|
|
|
+ &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}git; git-annex\\
|
|
|
+ (remote synchronization,\\
|
|
|
+ file availability and\\
|
|
|
+ integrity checks,\\
|
|
|
+ safe file deletion)\end{tabular} &
|
|
|
+ DOI registration
|
|
|
+ \\ \midrule
|
|
|
+ Findability &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}rich and standardized\\ metadata\end{tabular} &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}metadata aggregation\\
|
|
|
+ metadata search\end{tabular} &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}DOI registration;\\
|
|
|
+ DataCite support\\
|
|
|
+ repository search\end{tabular}
|
|
|
+ \\ \midrule
|
|
|
+Reproducibility &
|
|
|
+ &
|
|
|
+ \begin{tabular}[t]{@{}l@{}}run/rerun/container-run\\ functions\end{tabular} &
|
|
|
+
|
|
|
+\end{tabular}
|
|
|
+\caption{\textbf{\label{table:components}Contributions of each component of our proposed design in resolving the difficulties caused by daylong recordings} and laid out in Section \ref{section:problemspace}. ChildProject is a python package designed to perform recurrent tasks on the datasets; DataLad is a python package for the management of large, version-controlled datasets; GIN is a hosting provider dedicated to scientific data.}
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+\section{Proposed solution}
|
|
|
+
|
|
|
+
|
|
|
+\subsection{\label{sec:format}Dataset format}
|
|
|
+
|
|
|
+\begin{figure}[ht]
|
|
|
+ \centering
|
|
|
+ \inputTikZ{0.8}{Fig2.tex}
|
|
|
+ \caption{\textbf{Structure of a dataset}. Metadata, recordings and annotations each belong to their own folder. Raw annotations (i.e., the audio files as they have been collected, before post-processing) are separated from their post-processed counterparts (in this case: standardized and vetted recordings. Similarly, raw annotations -- in this case, LENA's its annotations -- are set apart from the corresponding CSV version.}
|
|
|
+ \label{fig:tree}
|
|
|
+\end{figure}
|
|
|
+
|
|
|
+To begin with, we propose a set of tested and proven standards which we use in our lab, and which build on previous experience in several collaborative projects, including ACLEW. It must be emphasized, however, that standards should be elaborated collaboratively by the community and that the following are merely a starting point.
|
|
|
+
|
|
|
+Data that are part of the same collection effort are bundled together within one folder\footnote{We believe a reasonable unit of bundling is the collection effort, for instance a single field trip, or a full bout of data collection for a cross-sectional sample, or a set of recordings done more or less at the same time in a longitudinal sample. Given the possibilities of versioning, some users may decide they want to keep all data from a longitudinal sample in the same dataset, adding to it progressively over months and years, to avoid having duplicate children.csv files. That said, given the system of subdatasets, one can always define different datasets, each of which contains the recordings collected in subsequent time periods.}, preferably a DataLad dataset (see Section \ref{section:datalad}). Datasets are packaged according to the structure given in fig. \ref{fig:tree}. The \path{metadata} folder contains at least three dataframes in CSV format : (i) \path{children.csv} contains information about the participants, such as their age or the language(s) they speak. (ii) \path{recordings.csv} contains the metadata for each recording, such as when the recording started, which device was used, or their relative path in the dataset. (iii) \path{annotations.csv} contains information concerning the annotations provided in the dataset, how they were produced, or which range they cover, etc. The dataframes are standardized according to guidelines which set conventional names for the columns and the range of allowed values. The guidelines are enforced through tests which print all the errors and inconsistencies in a dataset implemented in the ChildProject package introduced below.
|
|
|
+
|
|
|
+The \path{recordings} folder contains two subfolders: \path{raw}, which stores the recordings as delivered by the experimenters, and \path{converted} which contains processed copies of the recordings. All the audio files in \path{recordings/raw} are indexed in the recordings dataframe. There is, thus, no need for naming conventions for the audio files themselves, and maintainers can decide how they want to organize them.
|
|
|
+
|
|
|
+The \path{annotations} folder contains all sets of annotations. Each set itself consists of a folder containing two subfolders : i) \path{raw}, which stores the output of the annotation pipelines and ii) \path{converted}, which stores the annotations after being converted to a standardized CSV format and indexed into \path{metadata/annotations.csv}. A set of annotations can contain an unlimited amount of subsets, with any amount of recursions. For instance, a set of human-produced annotations could include one subset per annotator. Recursion facilitates the inheritance of access permissions, as explained in Section \ref{section:datalad}.
|
|
|
+
|
|
|
+
|
|
|
+\subsection{ChildProject}\label{section:childproject}
|
|
|
+
|
|
|
+The ChildProject package is a Python 3.6+ package that performs common operations on a dataset of child-centered recordings. It can be used from the command-line or by importing the modules from within Python. Assuming the target datasets are packaged according to the standards summarized in section \ref{sec:format}, the package supports the functions listed below.
|
|
|
+
|
|
|
+\subsubsection*{Listing errors and inconsistencies in a dataset}
|
|
|
+
|
|
|
+We provide a validation script that returns a detailed reporting of all the errors found within a dataset, such as violations of the formatting guidelines or missing files. Tests help enforce the standards that allow the commensurability of the datasets while guaranteeing the integrity and coherence of the data.
|
|
|
+
|
|
|
+\subsubsection*{Converting and indexing annotations}\label{section:annotations}
|
|
|
+
|
|
|
+The package converts input annotations to standardized, wide-table CSV dataframes. The columns in these wide-table formats have been determined based on previous work, and are largely specific to the goal of studying infants' language environment and production.
|
|
|
+
|
|
|
+Annotations are indexed into a unique CSV dataframe which stores their location in the dataset, the set of annotations they belong to, and the recording and time interval they cover. Thus, the index allows an easy retrieval of all the annotations that cover any given segment of audio, regardless of their original format and the naming conventions that were used. The system interfaces well with extant annotation standards. Currently, ChildProject supports: LENA annotations in .its \citep{xu2008lenatm}; ELAN annotations following the ACLEW DAS template \citep{Casillas2017,pympi-1.70}; the Voice Type Classifier (VTC) by \citet{lavechin2020opensource}; the Linguistic Unit Count Estimator (ALICE) by \citet{rasanen2020}; the VoCalisation Maturity Network (VCMNet) by \citet{AlFutaisi2019}. Users can also adapt routines for file types or conventions that vary; for instance, users can adapt the ELAN import developed for the ACLEW DAS template for their own template; and examples are also provided for Praat's .TextGrid files \citep{boersma2006praat}. The package also supports custom, user-defined conversion routines.
|
|
|
+
|
|
|
+Relying on the annotations index, the package can also calculate the intersection of the portions of audio covered by several annotators. This is useful when annotations from different annotators need to be combined (for instance, to retain the majority choice) or compared (e.g. for reliability evaluations).
|
|
|
+
|
|
|
+\subsubsection*{Choosing audio samples of the recordings to be annotated}\label{section:choosing}
|
|
|
+
|
|
|
+As noted in the Introduction, recordings are too extensive to be manually annotated in their entirety. We and colleagues have typically annotated manually clips of 0.5-5 minutes in length, and the way these clips are extracted and annotated constitutes one of the ways in which there is divergent standards (as illustrated in Table \ref{table:datasets}).
|
|
|
+
|
|
|
+The package allows the use of predefined or custom sampling algorithms. Samples' timestamps are exported to CSV dataframes. In order to keep track of the sample generating process, input parameters are simultaneously saved into a YAML file. Predefined samplers include a periodic sampler, a sampler targeting specific speakers' vocalizations, a sampler targeting regions of high-volubility according to input annotations, and a more agnostic sampler targeting high-energy regions. In all cases, the user can define the number of regions and their duration, as well as the context that may be inspected by human annotators. These options cover all documented sampling strategies.
|
|
|
+
|
|
|
+\subsubsection*{Generating ELAN files ready to be annotated}
|
|
|
+
|
|
|
+Although there was some variability in terms of the program used for human annotation, the field has now by and large settled on ELAN \citep{wittenburg2006elan}. ELAN employs xml files with hierarchical structure which are both customizable and flexible. The ChildProject can be used to generate .eaf files which can be annotated with the ELAN software based on samples of the recordings drawn using the package, as described in Section \ref{section:choosing}.
|
|
|
+
|
|
|
+\subsubsection*{Extracting and uploading audio samples to Zooniverse}
|
|
|
+
|
|
|
+The crowd-sourcing platform Zooniverse \citep{zooniverse} has been extensively employed in both natural \citep{gravityspy} and social sciences. More recently, researchers have been investigating its potential to classify samples of audio extracted from daylong recordings of children and the results have been encouraging \citep{semenzin2020a,semenzin2020b}. We provide tools interfacing with Zooniverse's API for preparing and uploading audio samples to the platform and for retrieving the result, while protecting the privacy of the participants.
|
|
|
+
|
|
|
+\subsubsection*{Audio processing}
|
|
|
+
|
|
|
+ChildProject allows the batch-conversion of the recordings to any target audio format (using ffmpeg \citealt{ffmpeg}).
|
|
|
+
|
|
|
+The package also implements a ``vetting" \citep{Cychosz2020} pipeline, which mutes segments of the recordings previously annotated by humans as confidential while preserving the duration of the audio files. After being processed, the recordings can safely be shared with other researchers or annotators.
|
|
|
+
|
|
|
+If necessary, users can easily design custom audio converters suiting more specific needs.
|
|
|
+
|
|
|
+\subsubsection*{Other functionalities}
|
|
|
+
|
|
|
+The package offers additional functions such as a pipeline that strips LENA's annotations from data that could be used to identify the participants, built upon previous code by \citet{eaf-anonymizer-original}.
|
|
|
+
|
|
|
+\subsubsection*{User empowerment}
|
|
|
+
|
|
|
+The present effort is led by a research lab, and thus with personnel and funding that is not permanent. We therefore have done our best to provide information to help the community adopt and maintain this code in the future. Extensive documentation is provided on \url{https://childproject.readthedocs.io}, including detailed tutorials. The code is accessible on GitHub.com.
|
|
|
+
|
|
|
+
|
|
|
+\subsection{DataLad}\label{section:datalad}
|
|
|
+
|
|
|
+\begin{figure}[htb]
|
|
|
+\centering
|
|
|
+\begin{minipage}{.5\linewidth}
|
|
|
+\centering
|
|
|
+\subfloat[]{\label{datalad:a}\resizebox{!}{0.70\linewidth}{\large\input{Fig1a.tex}\normalsize}}
|
|
|
+\end{minipage}%
|
|
|
+\begin{minipage}{.5\linewidth}
|
|
|
+\centering
|
|
|
+\subfloat[]{\label{datalad:b}\resizebox{!}{0.70\linewidth}{\large\input{Fig1b.tex}\normalsize}}
|
|
|
+\end{minipage}\par\medskip
|
|
|
+
|
|
|
+
|
|
|
+\caption{\label{fig:datalad}\textbf{DataLad development activity}. (a) Amount of versions published across time. More than 50 versions have been released since 2015-01-01, at a steady pace. (b) Share of commits held by top contributors in the last year (2020). At least three developers have contributed substantially, each of them being responsible for about 30\% of the commits.}
|
|
|
+
|
|
|
+\end{figure}
|
|
|
+
|
|
|
+The combination of standards and the ChildProject package allows us to solve some of the problems laid out in the Introduction, but they do not directly provide solutions to the problems of data sharing and collaborative work. DataLad, however, has been specifically designed to address such needs.
|
|
|
+
|
|
|
+DataLad \citep{datalad_handbook} was initially developed by researchers from the computational neuroscience community for the sharing of neuroimaging datasets. It has been under active development at a steady pace for at least six years (fig. \ref{datalad:a}). It is co-funded by the United States NSF and the German Federal Ministry of Education and Research and has several major code developers (fig. \ref{datalad:b}).% thereby lowering its bus-factor\footnote{\url{https://en.wikipedia.org/wiki/Bus_factor}} :D.
|
|
|
+
|
|
|
+DataLad relies on git-annex, a software designed to manage large files with git. Over the years, git has rapidly overcome competitors such as Subversion, and it has been popularized by platforms such as GitLab and GitHub. However, git does not natively handle large binary files, our recordings included. Git-annex circumvents this issue by versioning only pointers to the large files. The actual content of the files is stored in an ``annex''. Annexes can be stored remotely on a variety of supports, including Amazon Glacier, Amazon S3, Backblaze B2, Box.com, Dropbox, FTP/SFTP, Google Cloud Storage, Google Drive, Internet Archive via S3, Microsoft Azure Blob Storage, Microsoft OneDrive, OpenDrive, OwnCloud, SkyDrive, Usenet, and Yandex Disk.
|
|
|
+
|
|
|
+A DataLad dataset is, essentially, a git repository with an annex. As such, it naturally allows version control, easy collaboration with many contributors, and continuous testing. Furthermore, its use is intuitive to git users.
|
|
|
+
|
|
|
+In using git-annex, DataLad enables users to download only the files that they need, without needing to fetch the whole dataset.
|
|
|
+
|
|
|
+DataLad improves upon git-annex by adding a number of functionalities. One of them, dataset nesting, is built upon git submodules. A DataLad dataset can include sub-datasets, with as many levels of recursion as needed. This provides a natural solution to the question of how to document analyses, as an analysis repository can have the dataset on which it depends embedded as a subdataset. It also provides a good solution for the issue of different levels of data containing more or less identifying information, via the possibility of restricting permissions to different levels of the hierarchy.
|
|
|
+
|
|
|
+Like git, DataLad is a decentralized system, meaning that data can be stored and replicated across several ``remotes''. DataLad authors have argued in favor of decentralized research data management, as it simplifies infrastructure migrations, and help improve the scalibility of the data storage and distribution design \cite{decentralization_hanke}. Additionally, Decentralization is notably useful in that it helps achieve redundancy; files can be pushed simultaneously to several storage supports (e.g.: an external hard-drive, a cloud provider). In addition to that, when deleting large files from your local repository, DataLad will automatically make sure that more than a certain amount of remotes still own a copy the data, which by default is set to one.
|
|
|
+
|
|
|
+Many of the \emph{remotes} supported by DataLad require user-authentication, thus allowing for fine-grained access permissions management, such as Access-Control Lists (ACL). There are at least two ways to implement multiple levels of access within a dataset. One involves using sub-datasets with stricter access requirements. It is also possible to store data across several git-annex remotes with varying access permissions, depending on their sensitivity. Path-based pattern matching rules may configured in order to automatically select which remote the files should be pushed to. More flexible selection rules can be implemented using git-annex metadata, which allows to label files with \texttt{key=value} pairs. For instance, one could tag confidential files as \texttt{confidential=yes} and exclude these from certain remotes (blacklist). Alternatively, some files could be pushed to a certain remote provided they are labelled \texttt{public=yes} (whitelist).
|
|
|
+
|
|
|
+DataLad's metadata\footnote{\url{http://docs.datalad.org/en/stable/metadata.html}} system can extract and aggregate information describing the contents of a collection of datasets. A search function then allows the discovery of datasets based on these metadata. We have developed a DataLad extension to extract meaningful metadata from datasets into DataLad's metadata system \citep{datalad_extension}. This allows, for instance, to search for datasets containing a given language. Moreover, DataLad's metadata can natively incorporate DataCite \citep{brase2009datacite} descriptions into its own metadata.
|
|
|
+
|
|
|
+DataLad may link data and software dependencies associated to a script as it is run. These scripts can later be re-executed by others, and the dependencies will automatically be downloaded. This way, DataLad can keep track of how each intermediate file was generated, thus simplifying the reproducibility of analyses. DataLad's handbook provides a tutorial to create a fully reproducible paper \citep[Chapter~22]{datalad_handbook}, and a template is available on GitHub \citep{reproducible_paper}.
|
|
|
+
|
|
|
+DataLad is domain-agnostic, which makes it suitable for maturing techniques such as language acquisition studies based on long-form recordings. The open-access data of the WU-Minn Human Connectome Project \citep{pub.1022076283}, totalling 80 terabytes to date, have been made available through DataLad \footnote{\label{note:hcp}\url{https://github.com/datalad-datasets/human-connectome-project-openaccess}}.
|
|
|
+
|
|
|
+
|
|
|
+\subsection{Storage and distribution}\label{section:gin}
|
|
|
+
|
|
|
+DataLad does not provide, by itself, the infrastructure to share data. However, it allows maintainers to publish their content to a wide range of \href{https://git-annex.branchable.com/special_remotes/}{platforms}. One can therefore implement different strategies for the storage and distribution of the data using any combination of these providers, depending on the constraints.
|
|
|
+
|
|
|
+Table \ref{table:providers} sums up the most relevant characteristics of a few providers that are appropriate for our research, although many more could be considered. Datasets can only be cloned from providers that support git, and the large files can only be stored on those that support git-annex. Platforms that only support the former, such as GitHub, should therefore be used in tandem with providers that support the latter, like Amazon S3.
|
|
|
+
|
|
|
+Among criteria of special interest are: the provider's ability to handle complex permissions; how much data it can accept; its ability to assign permanent URLs and identifiers to the datasets; and of course, whether it complies with the legislation regarding privacy. For our purposes, Table \ref{table:providers} suggests GIN is the best option, handling well large files, with highly customizable permissions, and Git-based version control and access (see Appendix \ref{appendix:gin} for a practical use-case of GIN). That said, private projects are limited in space, although at the time of writing this limit can be raised by contributions to the GIN administrators. The next best option may be S3, and some users may prefer S3 when they do not have access to a local cluster, since S3 allows both easy storage and processing.
|
|
|
+
|
|
|
+To render comparison of options easier, detailed examples of storage designs taken from real datasets are listed in Appendix \ref{appendix:examples}. Scripts to implement these strategies can be found on our GitHub and OSF \citep{datalad_procedures}. We also provide a tutorial based on a public corpus \citep{vandam-day} to convert existing data to our standards and then publish it with DataLad\footnote{\url{https://childproject.readthedocs.io/en/latest/vandam.html}}.
|
|
|
+We would like to emphasize that the flexibility of DataLad makes it very easy to migrate from one architecture to another. The underlying infrastructure may change, with little to no impact on the users, and little efforts from the maintainers.
|
|
|
+
|
|
|
+In any case, we strongly recommend users to bear in mind that redundancy is important to make sure data are not lost, so a backup sibling may be hosted in an additional site (e.g., in a computer on campus in addition to the cloud-based version).
|
|
|
+
|
|
|
+For instance, \citet{Perkel_2019} suggests several practices regarding backups, including automated backups, privacy safe-guarding, regular tests, and offline backups. Table \ref{table:backups} may orient the reader towards the functionalities of DataLad (and git-annex) which can be used to achieve these goals.
|
|
|
+
|
|
|
+\begin{table*}[ht]
|
|
|
+
|
|
|
+\begin{minipage}{\columnwidth}%
|
|
|
+\centering
|
|
|
+\renewcommand\footnoterule{ \kern -1ex}
|
|
|
+\renewcommand{\thempfootnote}{\alph{mpfootnote}}
|
|
|
+
|
|
|
+\begin{tabular}{@{}lllllll@{}}
|
|
|
+\toprule
|
|
|
+\multicolumn{1}{c}{\textbf{Provider}} & \multicolumn{1}{c}{\textbf{Git}\footnote{The provider can store the git history and provide an URL from which the dataset can be installed.}} & \multicolumn{1}{c}{\textbf{Large files}\footnote{The provider handles git-annex large files.}} & \multicolumn{1}{c}{\textbf{Authentication}} & \multicolumn{1}{c}{\textbf{Permissions}} & \multicolumn{1}{c}{\textbf{Quota}} &
|
|
|
+\multicolumn{1}{c}{\textbf{\begin{tabular}[t]{@{}l@{}}DOI\\registration\end{tabular}}} \\ \midrule
|
|
|
+\midrule
|
|
|
+SSH server & Yes & Yes & SSH & Unix & Self-hosted & No \\
|
|
|
+GIN & Yes & Yes & HTTPS or SSH & ACL & \footnote{\label{contact}Contact the administrators}
|
|
|
+ & Yes\footnoteref{contact}\\
|
|
|
+GitHub & Yes & No & HTTPS or SSH & ACL & -- & No \\
|
|
|
+GitLab & Yes & No & HTTPS or SSH & ACL & -- & No \\
|
|
|
+
|
|
|
+Nextcloud & No & Yes & & ACL & Self-hosted & No \\
|
|
|
+Amazon S3 & No & Yes & API key+secret & IAM & Unlimited & No \\
|
|
|
+OSF & Yes\footnote{\label{osf}With limitations (see \url{http://docs.datalad.org/projects/osf/en/latest/intro.html})} & Yes\footnoteref{osf} & Token & ACL & \footnote{5 GB for private projects, 50 GB for publics projects} & Yes \\
|
|
|
+\end{tabular}
|
|
|
+\end{minipage}
|
|
|
+\caption{\label{table:providers}\textbf{Overview of several providers that can be used with DataLad}. The Unix permission system allows only one user and one group to be granted specific access rights. Access Control Lists (ACL) give more control, by enabling access to several groups and users. Amazon's Identity Access Management (IAM) can imitate ACLs, while providing more functionalities (fully-programmable; time-limited permissions; etc.) }
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+\begin{table*}[ht]
|
|
|
+\begin{minipage}{\columnwidth}%
|
|
|
+\centering
|
|
|
+\renewcommand\footnoterule{ \kern -1ex}
|
|
|
+\renewcommand{\thempfootnote}{\alph{mpfootnote}}
|
|
|
+\begin{tabular}{@{}lll@{}}
|
|
|
+\toprule
|
|
|
+Practice & \begin{tabular}[t]{@{}l@{}}Relevant\\ software\end{tabular} & Functionality \\ \midrule\midrule
|
|
|
+\begin{tabular}[t]{@{}l@{}}offline\\ backups\end{tabular} & \begin{tabular}[t]{@{}l@{}}DataLad\\ \ \\ git-annex\end{tabular} & \begin{tabular}[t]{@{}l@{}}create-sibling, push\footnote{creates a local sibling to which the data can be pushed, e.g. an external hard-drive.};\\export-archive;\\copy;export\footnote{exports human-readable snapshots of a dataset}\end{tabular} \\ \midrule
|
|
|
+\begin{tabular}[t]{@{}l@{}}backup\\ automation\end{tabular} & DataLad & \begin{tabular}[t]{@{}l@{}}siblings\\ ``publish-depends''\footnote{``publish-depends'' specifies which other siblings should be pushed to everytime some other sibling is updated. Maintainers can thus make sure that pushing to the main repository will trigger a push to the backup sibling.}\end{tabular} \\ \midrule
|
|
|
+\begin{tabular}[t]{@{}l@{}}privacy\\ safe-guarding\end{tabular} & git-annex & encryption \\ \midrule
|
|
|
+regular tests & git-annex & \texttt{fsck}\footnote{integrity check} \\ \bottomrule
|
|
|
+\end{tabular}
|
|
|
+\caption{\label{table:backups}\textbf{Examples of recommended practices for data backups, associated to the software that could be used for their implementation}.}
|
|
|
+\end{minipage}
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+\section{Application: evaluating annotations' reliability}
|
|
|
+
|
|
|
+
|
|
|
+Assessing the reliability of the annotations is crucial to linguistic research, but it can prove tedious in the case of daylong recordings. On one hand, analysis of the massive amounts of annotations generated by automatic tools may be computationally intensive. On the other hand, human annotations are usually sparse and thus more difficult to match with each other. Moreover, as emphasized in Section \ref{section:problemspace}, the variety of file formats used to store the annotations makes it even harder to compare them.
|
|
|
+
|
|
|
+Making use of the consistent data structures that it provides, the ChildProject package implements functions for extracting and aligning annotations regardless of their provenance or nature (human vs algorithm, ELAN vs Praat, etc.). It also provides functions to compute most of the metrics commonly used in linguistics and speech processing, relying on existing efficient and field-tested implementations.
|
|
|
+
|
|
|
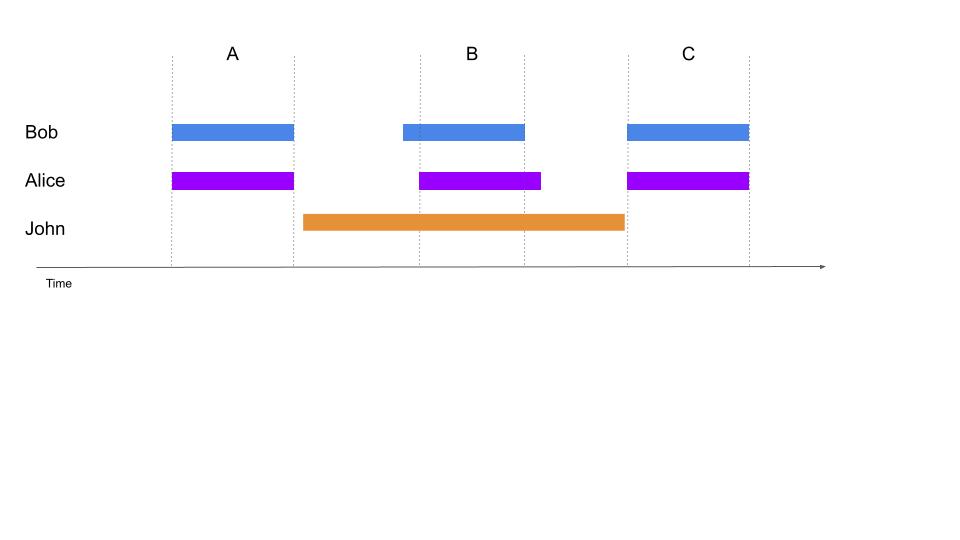
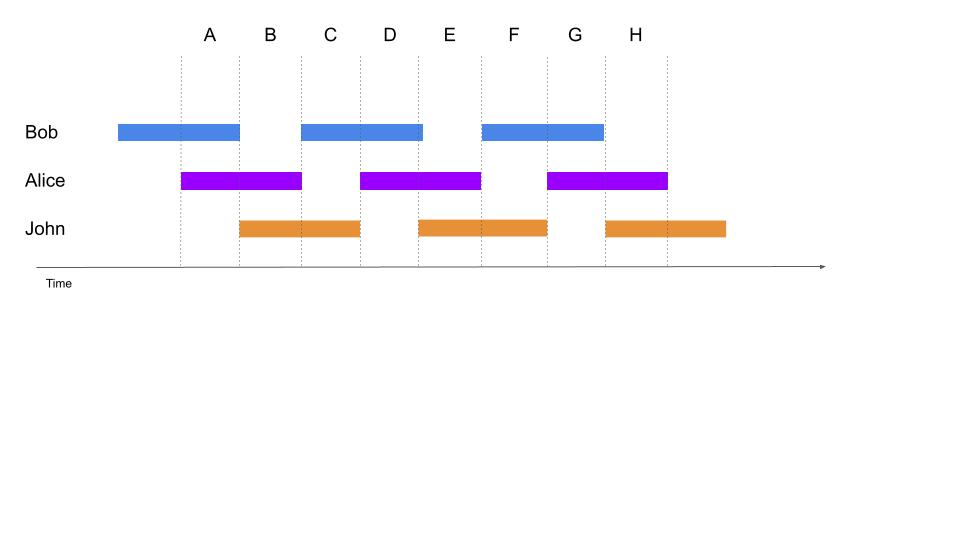
+Figure \ref{fig:Annotation} illustrates a recording annotated by three annotators (Alice, Bob and John). In this case, if one is interested in comparing the annotations of Bob and Alice, then the segments A, B and C should be compared. However, if the annotations common to all of the three annotators should be simultaneously compared, only the segment B should be considered.
|
|
|
+In real datasets with many recordings and several human and automatic annotators, the layout of annotations coverage may become unpredictable. Relying on the index of annotations described in Section \ref{section:annotations}, the ChildProject package can calculate the intersection of the portions of audio covered by several annotators and return all matching annotations. These annotations can be filtered (e.g. excluding certain audio files), grouped according to certain characteristics (e.g. by participant), or collapsed altogether for subsequent analyses.
|
|
|
+
|
|
|
+
|
|
|
+\begin{figure*}[htb]
|
|
|
+\centering
|
|
|
+\subfloat[]{%
|
|
|
+\centering
|
|
|
+ \includegraphics[trim=0 250 100 25, clip, width=0.8\textwidth]{Fig3a.jpg}
|
|
|
+ \label{Annotation:1}%
|
|
|
+}
|
|
|
+
|
|
|
+%\subfloat[]{%
|
|
|
+%\centering
|
|
|
+% \includegraphics[trim=0 250 25 25, clip, width=0.98\textwidth]{Fig3b.jpg}
|
|
|
+% \label{Annotation:2}%
|
|
|
+%}
|
|
|
+
|
|
|
+\caption{\label{fig:Annotation}\textbf{Example of time-intervals of a recording covered by three annotators}. Automated annotations usually cover whole recordings, while human annotators typically annotate periodic or targeted clips. }
|
|
|
+
|
|
|
+\end{figure*}
|
|
|
+
|
|
|
+
|
|
|
+In psychometrics, the reliability of annotators is usually evaluated using inter-coder agreement indicators. The python package enables the calculation of some of these measures. Krippendorff's Alpha and Fleiss' Kappa \citep{kappa} have been implemented with NLTK \citep{nltk}. The gamma method \citep{gamma}, which aims to improve upon previous indicators by evaluating simultaneously the quality of both the segmentation and the categorization of speech, has been included using the implementation by \citet{pygamma_agreement}.
|
|
|
+It should be noted that these measures are most useful in the absence of ground truth, when reliability of the annotations can only be assessed by evaluating their overall agreement. Automatic annotators, however, are usually evaluated against a gold standard produced by human experts. In such cases, the package allows comparisons of pairs of annotators using metrics such as F-score, recall, and precision. Figure \ref{fig:precision} illustrates this functionality. Additionally, the package can compute confusion matrices between two annotators, allowing more informative comparisons, as demonstrated in Figure \ref{fig:confusion}. Finally, the python package interfaces well with \texttt{pyannote.metrics} \citep{pyannote.metrics}, and all the metrics implemented by the latter can be effectively used on the annotations managed with ChildProject.
|
|
|
+
|
|
|
+\begin{figure*}[htb]
|
|
|
+
|
|
|
+\centering
|
|
|
+\includegraphics[width=0.8\textwidth]{Fig4.pdf}
|
|
|
+
|
|
|
+\caption{\label{fig:precision}\textbf{Examples of diarization performance evaluation using recall, precision and F1 score}. Audio from the the public VanDam corpus \citep{vandam-day} is annotated according to who-speak-when, using both the LENA diarizer (its) and the Voice Type Classifier (VTC) by \citet{lavechin2020opensource}. Speech segments are classified among four speaker types: the key child (CHI), other children (OCH), male adults (MAL) and female adults (FEM). For illustration purposes, fake annotations are generated from that of the VTC. Two are computed by randomly assigning the speaker type to 50\% and 75\% (conf) of the VTC's speech segments. Two are computed by dropping 50\% of speech segments from the VTC (drop). Recall, precision and F1 score are calculated for each of these annotations, by comparing them to the VTC. The worst F-score for the LENA is reached for OCH segments. Dropping segments does not alter precision; however, as expected, it has a substantially negative impact on recall.
|
|
|
+}
|
|
|
+
|
|
|
+\end{figure*}
|
|
|
+
|
|
|
+
|
|
|
+\begin{figure*}[htb]
|
|
|
+
|
|
|
+\centering
|
|
|
+\includegraphics[width=\textwidth]{Fig5.pdf}
|
|
|
+
|
|
|
+\caption{\label{fig:confusion}\textbf{Example of diarization performance comparison using confusion matrices}
|
|
|
+LENA's annotations (its) of the public VanDam corpus \citep{vandam-day} are compared to the VTC's. The first coefficient of the left side matrix should be read as: ``41\% of CHI segments from the VTC are labelled as CHI by the LENA's''. The first coefficient of the right side matrix should be read as: ``71\% of the CHI segments of the LENA are labelled as CHI by the VTC''. It can also be seen that the LENA does not produce overlapping speech segments, i.e. it cannot disambiguate two overlapping speakers.
|
|
|
+}
|
|
|
+
|
|
|
+\end{figure*}
|
|
|
+
|
|
|
+
|
|
|
+%\subsubsection{Possible improvements}
|
|
|
+% adding pyanote
|
|
|
+% pygamma
|
|
|
+% generalizing to other annotation types
|
|
|
+
|
|
|
+
|
|
|
+\section{Generalization}
|
|
|
+
|
|
|
+The kinds of problems that our proposed approach addresses are relevant to at least three other bodies of data, all of them based on large datasets collected with wearables. First, there is a line of research on interaction and its effects on well-being among neurotypical adults (e.g., \cite{ear1}). Second, audio data from wearables holds promise for individuals with medical and psychological conditions that have behavioral consequences which can evolve over time, including conditions that lead to coughing \citep{Wu2018} and/or neurogenerative disorders \citep{riad2020vocal}. Third, some researchers hope to gather datasets on child development combining multiple information sources, such as parental reports, as well as other sensors picking up motion and psychophysiological data, with the goal of potentially intervening when it is needed \citep{levin2021sensing}.
|
|
|
+
|
|
|
+Our proposed solution can be readily adapted to the first body of data: All that would need to be changed is renaming children.csv to participants.csv; renaming child\_id to participant\_id; and adapting which columns are mandatory and their format (e.g., it is cumbersome to express age in days for adults).
|
|
|
+
|
|
|
+Generalizing our solution to the second body of data requires more adaptation. For such use cases, it would be ideal for the equipment to be left in the patients' house, so that it can be used for instance one day a week or month. Additional work is needed to facilitate this, ranging from making the equipment easier to use and more robust by for instance facilitating charging and secure data transfer from such off-site locations.
|
|
|
+
|
|
|
+The third use case requires further adaptation, in addition to those just mentioned (making the sensors easy to use and allowing data transfer from potentially insecure home settings). In particular, multiple sensors' data need to be integrated together and time-stamped. We have made some progress in this sense in the context of the collection of multiple audio tracks collected with different physical devices (example on XXX), but have not yet developed structure and code to support the integration of pictures, videos, heart rate data, parental questionnaire data, etc.
|
|
|
+
|
|
|
+\section{Limitations}
|
|
|
+
|
|
|
+DataLad and git-annex are well-documented, and, on the user's end, little knowledge beyond that of git is needed. Maintainers and resource administrators, however, will need a certain level of understanding in order to take full advantage of these tools.
|
|
|
+Recently, \citet{Powell2021} has emphasized the shortcomings of decentralization and the inconveniences of a proliferation of databases with different access protocols. In the future, sharing data could be made even easier if off-the-shelf solutions compatible with DataLad were made readily available to linguists, psychologists, and developmental scientists. To this effect, we especially call for the attention of our colleagues working on linguistic databases. We are pleased to have found a solution on GIN -- but it is possible that GIN administrators agreed to host our data because there is some potential connection with neuroimaging, whereas they may not be able to justify their use of resources for under-resourced languages and/or other projects that bear little connection to neuroimaging.
|
|
|
+
|
|
|
+We should stress again that the use of the ChildProject package does not require the datasets to be managed with DataLad. They do need, however, to follow certain standards. Standards, of course, do not come without their own issues, especially in the present case of a maturing technique. They may be challenged by ever-evolving software, hardware, and practices. However, we believe that the benefits of standardization outweigh its costs provided that it remains reasonably flexible. Such standards will further help to combine efforts from different teams across institutions. More procedures and scripts that solve recurrent tasks can be integrated into the ChildProject package, which might also speed up the development of future tools.
|
|
|
+One could argue that new standards usually most usually end up increasing the amount of competing standards instead of bringing consensus. Nonetheless, if one were to eventually impose itself, well-structured datasets would still be easier to adapt than disordered data representations. Meanwhile, we look forward to discussing standards collaboratively with other teams via the GitHub platform, where anyone can create issues for improvements or bugs, submit pull-requests to integrate an improvement they have made, and/or have relevant conversations in the forum.
|
|
|
+
|
|
|
+\section{Summary}
|
|
|
+
|
|
|
+% removed: assessing data reliability
|
|
|
+% Data managers should be interested in DataLad because it might benefit to many studies, beyond long-form recordings. We should convince them it is worth diving into it
|
|
|
+
|
|
|
+We provide a solution to the technical challenges related to the management, storage and sharing of datasets of child-centered day-long recordings. This solution relies on four components: i) a set of standards for the structuring of the datasets; ii) \emph{ChildProject}, a python package to enforce these standards and perform useful operations on the datasets; iii) DataLad, a mature and actively developed version-control software for the management of scientific datasets; and iv) GIN, a storage provider compatible with Datalad. Building upon these standards, we have also provide tools to simplify the extraction of information from the annotations and the evaluation of their reliability along with the python package. The four components of our proposed design serve partially independent goals and can thus be decoupled, but we believe their combination would greatly benefit the technique of long-form recordings applied to language acquisition studies.
|
|
|
+
|
|
|
+\section*{Declarations}
|
|
|
+
|
|
|
+\subsubsection*{Funding}
|
|
|
+This work has benefited from funding and/or institutional support from Agence Nationale de la Recherche (ANR-17-CE28-0007 LangAge,
|
|
|
+ANR-16-DATA-0004 ACLEW, ANR-14-CE30-0003 MechELex, ANR-17-EURE-0017);
|
|
|
+and the J. S. McDonnell Foundation Understanding Human Cognition Scholar Award. We also benefited from code developed in the Bridges system, which is
|
|
|
+supported by NSF award number ACI-1445606, at the Pittsburgh
|
|
|
+Supercomputing Center (PSC), using the Extreme Science and Engineering Discovery Environment
|
|
|
+(XSEDE), which is supported by National Science Foundation grant number OCI-1053575. Additionally, we benefited from processing in GENCI-IDRIS, France (Grant-A0071011046). Some capabilities of our software depend on the Zooniverse.org platform, the development of which is funded by generous support, including a Global Impact Award from Google, and by a grant from the Alfred P. Sloan Foundation. The funders had no impact on this study.
|
|
|
+
|
|
|
+\subsubsection*{Conflicts of interest/Competing interests}
|
|
|
+
|
|
|
+The authors have no conflict of interests to disclose.
|
|
|
+
|
|
|
+\subsubsection*{Availability of data and material}
|
|
|
+This paper does not directly rely on specific data or material.
|
|
|
+
|
|
|
+\subsubsection*{Code availability}
|
|
|
+
|
|
|
+The ChildProject package is available on GitHub at \url{https://github.com/LAAC-LSCP/ChildProject}. We provide scripts and templates for DataLad managed datasets at \url{http://doi.org/10.17605/OSF.IO/6VCXK} \citep{datalad_procedures}. We also provide a DataLad extension to extract metadata from corpora of daylong recordings \citep{datalad_extension}.
|
|
|
+Examples of annotations evaluations using the package can be found at XXX.
|
|
|
+
|
|
|
+\appendix
|
|
|
+
|
|
|
+\section{Examples of storage strategies}\label{appendix:examples}
|
|
|
+
|
|
|
+\subsection{\label{sec:example1}Example 1 - sharing a dataset within the lab}
|
|
|
+
|
|
|
+In the first example, Alice is hosting large datasets of a few terabytes of recordings and annotations and she wants to share them with Bob - a collaborator from her own institution - in a secure manner. Alice and Bob are familiar with GitHub, and they like its user-friendly features such as issues and pull requests. However, GitHub cannot handle such amounts of data.
|
|
|
+
|
|
|
+Alice decides to store the git repository itself on GitHub -- or a GitLab instance, it would not matter -- thus allowing to benefit from their nice features while hosting the large files -- the recordings and annotations -- elsewhere. Alice's laboratory has its own cluster, with a large storage capacity. Thus, she decides to host the files there for free rather than using a Cloud provider.
|
|
|
+
|
|
|
+Since Bob has been given SSH access to the cluster and belongs to the right UNIX group, he can download recordings and annotations from their joint institution cluster. Alice also made sure to configure the dataset in a way that makes sure every change published to GitHub is also published to the cluster, with DataLad's ``publish-depends'' option.
|
|
|
+
|
|
|
+For backup purposes, a third sibling is hosted on Amazon S3 Glacier -- which is cheaper than S3 at the expense of higher retrieval costs and delays -- as a git-annex \href{https://git-annex.branchable.com/special_remotes/}{special remote}. Special remotes do not store the git history and they cannot be used to clone the dataset. However, they can be used as a storage support for the recordings and other large files. In order to increase the security of the data, Alice uses encryption. Git-annex implements several encryption schemes\footnote{\url{https://git-annex.branchable.com/encryption/}}. The hybrid scheme allows to add public GPG keys at any time without additional decryption/encryption steps. Each user can then later decrypt the data with their own private key. This way, as long as at least one private GPG key has not been lost, data are still recoverable. This is especially valuable in that in naturally ensures redundancy of the decryption keys, which is critical in the case of encrypted backups.
|
|
|
+
|
|
|
+By default, file names are hashed with an HMAC algorithm, and their content is encrypted with AES-128 -- GPG's default --, although another algorithm could be selected.
|
|
|
+
|
|
|
+This setup ensures redundancy of git files (hosted on both GitHub and the cluster) as well as large files (stored on both the cluster and Amazon Deep Glacier). It also allows Bob to signal and correct errors he finds, and/or to add annotations in a straightforward manner, benefiting Alice. By virtue of having siblings, they can make sure that their local dataset is organized in an identical manner, facilitating collaboration and reproducibility in their analyses.
|
|
|
+
|
|
|
+Table \ref{table:storage1} illustrates such a strategy. In this example, users install the dataset from a private GitHub repository. Continuous testing is configured with Travis CI\footnote{https://travis-ci.com/}, in order to ensure the integrity of the dataset at every step. GitHub Actions could also be used for that purpose\footnote{https://docs.github.com/en/actions}.
|
|
|
+
|
|
|
+We used this strategy -- minus the Glacier backups -- to maintain and deliver 4 datasets with 8700 hours of audio\footnote{\url{https://github.com/LAAC-LSCP/datasets}} for several months. The associated scripts can be found on \cite{datalad_procedures}. We have now transitioned to using GIN for our main site, with our cluster as the backup. The scripts associated to this set-up can be found at the same location.
|
|
|
+
|
|
|
+
|
|
|
+\begin{table*}[!htbp]
|
|
|
+\centering
|
|
|
+\begin{tabular}{@{}lllllll@{}}
|
|
|
+\toprule
|
|
|
+\textbf{Sibling} & \textbf{Provider} & \textbf{Content} & \textbf{Access} & \textbf{Encryption} \\ \midrule
|
|
|
+origin & GitHub & metadata; scripts & Lab & No \\
|
|
|
+cluster & SSH server & everything & Lab & No \\
|
|
|
+backup & Amazon Deep Glacier & recordings; annotations & Lab & AES-128 \\ \bottomrule
|
|
|
+\end{tabular}
|
|
|
+\caption{\label{table:storage1}Example 1 - Storage strategy example relying on GitHub and a cluster to deliver the data.}
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+\subsection{Example 2 - sharing large datasets with outside collaborators (S3)}
|
|
|
+
|
|
|
+The previous strategy is not suitable when complex permissions are required, since SSH remotes only handle Unix-style permissions (user, group, all).
|
|
|
+
|
|
|
+Moreover, Alice may want to share the dataset with collaborators outside her lab, without giving them SSH access to its cluster. Or, she may not even own the infrastructure that would allow her to store and share such large amounts of data.
|
|
|
+
|
|
|
+Instead, she decides to use Amazon S3 together with GitHub. Authorized users are provided their own Amazon S3 API key and secret, which are managed with Amazon's Identity and Access Manager (IAM). The GitHub is stripped from all confidential data, which are stored in the S3 annex only, allowing to manage access permissions entirely through IAM. This strategy is used by the Human Connectome Project\footnoteref{note:hcp}.
|
|
|
+
|
|
|
+Furthermore, Alice makes sure to encrypt GDPR relevant data, using strong symmetric encryption (AES-128). This strategy is illustrated in Table \ref{table:storage2}.
|
|
|
+
|
|
|
+\begin{table*}[!htbp]
|
|
|
+\centering
|
|
|
+\begin{tabular}{@{}lllllll@{}}
|
|
|
+\toprule
|
|
|
+\textbf{Sibling} & \textbf{Provider} & \textbf{Content} & \textbf{Access} & \textbf{Encryption} \\ \midrule
|
|
|
+origin & GitHub & metadata; scripts & Collaborators & No \\
|
|
|
+s3 & Amazon S3 & recordings; annotations & Collaborators & AES-128 \\ \bottomrule
|
|
|
+\end{tabular}
|
|
|
+\caption{\label{table:storage2}Example 2 - Storage strategy example relying on GitHub and Amazon S3.}
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+Amazon is superior to most alternatives for a number of reasons, including that it is highly tested, developed by engineers with a high-level of knowledge of the platform, and widely used. This means that the code is robust even before it is released, and it is widely tested once it is released. The fact that there are many users also entails that issues or questions can be looked up online. In addition, in the context of data durability, Amazon is a good choice because it is too big to fail, and thus probably available for the long-term. In addition, in sheer terms of flexibility and coverage, Amazon provides a whole suite of tools (for data sharing, backups, and processing), which may be useful for researchers with little access to high-capacity infrastructures.
|
|
|
+
|
|
|
+\subsection{Example 3 - sharing large datasets with outside collaborators and multi-tier access (GIN)}\label{appendix:gin}
|
|
|
+
|
|
|
+Due to legislation in some countries, there are researchers who may not be authorized to store their data on Amazon. If they also do not have access to a local cluster (see Example 1) and/or even in the case that they have a local cluster, but need finer control of access permissions, there are alternatives which can be used as a workaround.
|
|
|
+
|
|
|
+Finding herself in this setting, Alice decides to use the G-Node Infrastructure (GIN)\footnote{\url{https://gin.g-node.org/}}, which is dedicated to providing ``Modern Research Data Management for Neuroscience''. GIN is similar to GitLab and GitHub in many aspects, except that is also supports git-annex and thus can directly host the large files that required third-party providers while using those platforms.
|
|
|
+
|
|
|
+Just like GitLab or GitHub, it can handle complex permissions, at the user or group-level, thus surpassing Unix-style permissions management.
|
|
|
+
|
|
|
+In this case, Alice needs three permission tiers: 1) read-only access to anonymized data, 2) read-only access to confidential data, and 3) read and write access to the whole data. In order to achieve this, she creates two GIN siblings per dataset: \texttt{origin} and \texttt{confidential}. The dataset is configured to publish all the files whose path contains \path{/confidential/} to the \path{confidential} repository, while the rest of the data is published to \texttt{origin}. Alice could then great read-only access to \texttt{origin} to both Bob and Carol, while restricting the access to \texttt{confidential} to Bob only.
|
|
|
+
|
|
|
+Since Alice has not been allowed to use a cloud provider, and is lacking a local infrastructure, she needs an alternate solution for her backups. She may use external hard drives, as DataLad allows to push data to a local storage as with any other kind of storage.
|
|
|
+
|
|
|
+Table \ref{table:storage3} sums up this strategy, which is currently used to deliver the EL1000 dataset\footnote{\url{https://gin.g-node.org/EL1000/EL1000}} -- except for the backup, which is located at our cluster --. The EL1000 is a composite dataset, created by the contribution of 18 different teams that collected data independently but using comparable methods.
|
|
|
+
|
|
|
+\begin{table*}[!htbp]
|
|
|
+\centering
|
|
|
+\begin{tabular}{@{}lllllll@{}}
|
|
|
+\toprule
|
|
|
+\textbf{Sibling} & \textbf{Provider} & \textbf{Content} & \textbf{Access} & \textbf{Encryption} \\ \midrule
|
|
|
+origin & GIN & files NOT matching \path{**/confidential/*} & \begin{tabular}[t]{@{}l@{}}Alice (read+write);\\Bob, Carol (read-only)\end{tabular} & No \\
|
|
|
+confidential & GIN & files matching \path{**/confidential/*} & \begin{tabular}[t]{@{}l@{}}Alice (read+write);\\Bob (read-only)\end{tabular} & No \\
|
|
|
+backup & \begin{tabular}[t]{@{}l@{}}external\\hard drive\end{tabular} & everything & Alice & No \\\bottomrule
|
|
|
+\end{tabular}
|
|
|
+\caption{\label{table:storage3}Example 3 - Storage strategy example relying solely on GIN to deliver the data.}
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+
|
|
|
+\subsection{Example 4 - Sharing smaller datasets (OSF)}
|
|
|
+
|
|
|
+
|
|
|
+The Open Science Framework (OSF) is especially interesting because it supports DOI registration, providing permanent URLs to access the datasets. Moreover, an extension of DataLad has specifically been developed to work with OSF, which may host both the git repository and the large files (see Table \ref{table:providers}). In addition, Shibboleth credentials can be used with OSF.
|
|
|
+
|
|
|
+Low quotas are an important downside with OSF. Public projects are limited to 50 GB, and private projects cannot exceed 5 GB, which is too low for most long-form datasets.
|
|
|
+However, OSF could be used only to host the git repository, effectively providing a permanent URL from which the dataset can be installed, as long as the content of the large files remains available from a third-party provider, e.g. with Amazon S3. Table \ref{table:storage4} illustrates such a strategy.
|
|
|
+
|
|
|
+\begin{table*}[!htbp]
|
|
|
+\centering
|
|
|
+\begin{tabular}{@{}lllllll@{}}
|
|
|
+\toprule
|
|
|
+\textbf{Sibling} & \textbf{Provider} & \textbf{Content} & \textbf{Access} & \textbf{Encryption} \\ \midrule
|
|
|
+origin & OSF & metadata; scripts & Everyone & No \\
|
|
|
+s3 & Amazon S3 & annotations; recordings & Alice, Bob and Carol & No \\ \bottomrule
|
|
|
+\end{tabular}
|
|
|
+\caption{\label{table:storage4}Example 4 - Storage strategy example relying on OSF and Amazon S3 to deliver the data.}
|
|
|
+\end{table*}
|
|
|
+
|
|
|
+We use a reversed approach for our demo dataset\footnote{https://github.com/LAAC-LSCP/vandam-daylong-demo} based on \citep{vandam-day}, by hosting the git repository on GitHub, and hosting the large files on OSF. This is possible only because of the small size of the dataset.
|
|
|
+
|
|
|
+\bibliographystyle{spbasic}
|
|
|
+\bibliography{references}
|
|
|
+
|
|
|
+\end{document}
|