| | \n",
" | \n",
"
Introduction to\n", "\n", "
Representing electrophysiology data in Python"
]
},
{
"cell_type": "markdown",
"id": "d02e9275",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"## Data sources in electrophysiology\n",
"\n",
"\n"
]
},
{
"cell_type": "markdown",
"id": "dee2a771",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Data modalities in electrophysiology\n",
"\n",
"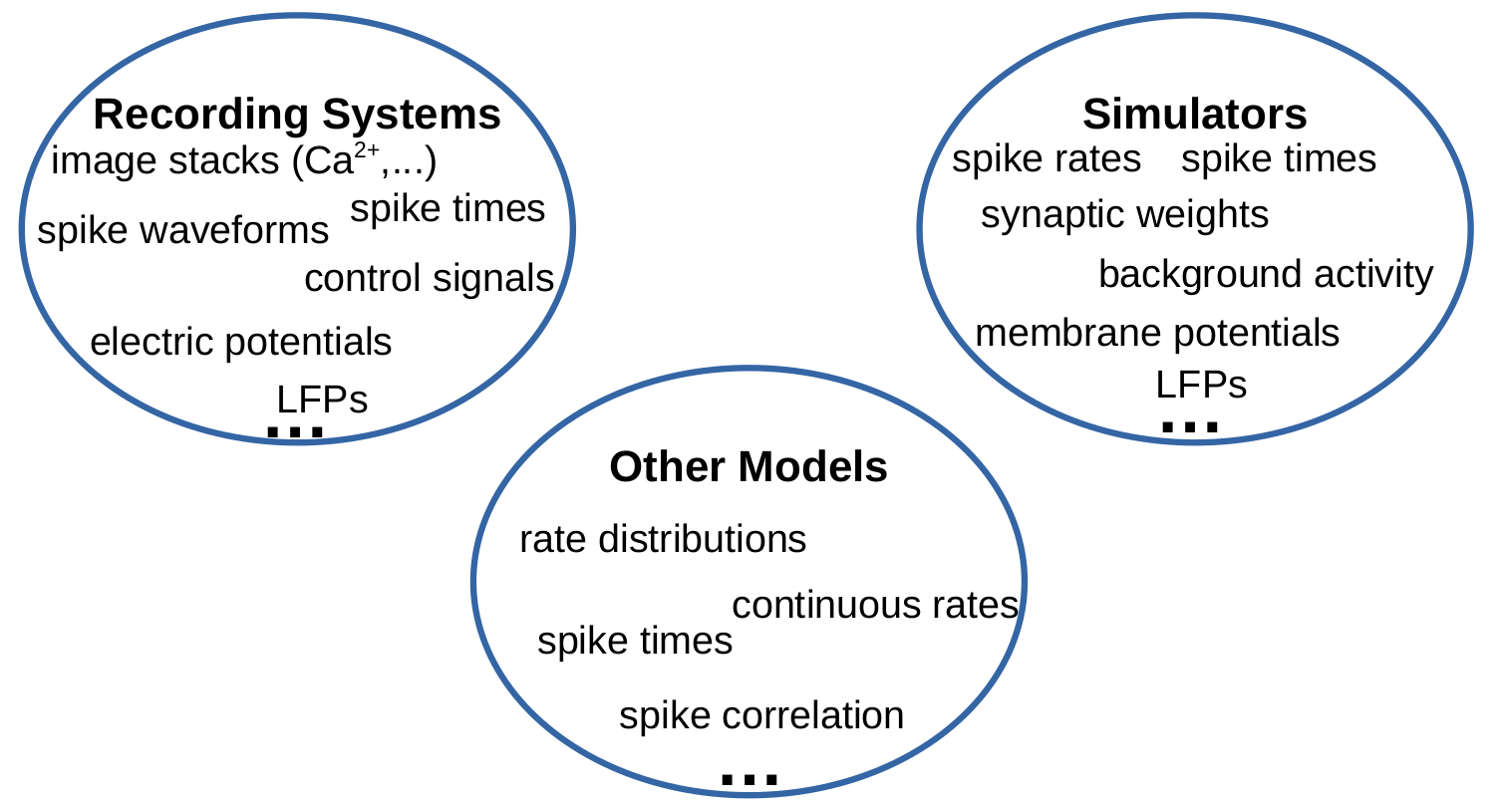\n"
]
},
{
"cell_type": "markdown",
"id": "4c7b4932",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"How can these diverse data types and formats by treated in a common framework to \n",
"- allow combined analyses\n",
"- facilitate reproducibility\n",
"- simplify scientific workflows"
]
},
{
"cell_type": "markdown",
"id": "b80bdd60",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"source": [
"...without inventing yet another file format standard?"
]
},
{
"cell_type": "markdown",
"id": "c09c4aac",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"## Neo - Generic representation of common ephys modalities\n",
"- Standardized representation of ephys data during runtime\n",
"- Efficient handling of large data arrays thanks to `numpy`\n",
"- Description of physical units and unit conversion thanks to `quantities`"
]
},
{
"cell_type": "code",
"execution_count": 19,
"id": "6e5df85c",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [],
"source": [
"import neo\n",
"import numpy as np\n",
"import quantities as pq"
]
},
{
"cell_type": "markdown",
"id": "9242b2de",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Data Classes\n",
"- `AnalogSignal`: Continuous data sampled in **regular** intervals"
]
},
{
"cell_type": "markdown",
"id": "7f8c47d0",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"source": [
"*essential metadata*: physical unit of samples, time stamps of the samples (first timestamp & sampling interval)"
]
},
{
"cell_type": "code",
"execution_count": 34,
"id": "ecfbe500",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [],
"source": [
"anasig = neo.AnalogSignal(np.zeros((50,2)), units='uV',\n",
" sampling_rate=10000*pq.Hz,\n",
" t_start=120*pq.ms)"
]
},
{
"cell_type": "markdown",
"id": "fa04abd6",
"metadata": {
"slideshow": {
"slide_type": "-"
}
},
"source": [
"Note the dimensions of `Analogsignal` object: (`time`, `channel`)"
]
},
{
"cell_type": "markdown",
"id": "710a0eb7",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"#### Accessing metadata"
]
},
{
"cell_type": "code",
"execution_count": 37,
"id": "9ba68d73",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"units: 1.0 uV\n",
"sampling_rate: 10000.0 Hz\n",
"sampling_period: 0.0001 s\n",
"t_start & t_stop: (array(120.) * ms, array(125.) * ms)\n",
"times: [120. 120.1 120.2 120.3 120.4 120.5 120.6 120.7 120.8 120.9 121. 121.1\n",
" 121.2 121.3 121.4 121.5 121.6 121.7 121.8 121.9 122. 122.1 122.2 122.3\n",
" 122.4 122.5 122.6 122.7 122.8 122.9 123. 123.1 123.2 123.3 123.4 123.5\n",
" 123.6 123.7 123.8 123.9 124. 124.1 124.2 124.3 124.4 124.5 124.6 124.7\n",
" 124.8 124.9] ms\n"
]
}
],
"source": [
"print(f'units: {anasig.units}')\n",
"print(f'sampling_rate: {anasig.sampling_rate}')\n",
"print(f'sampling_period: {anasig.sampling_period.simplified}')\n",
"print(f't_start & t_stop: {anasig.t_start, anasig.t_stop}')\n",
"print(f'times: {anasig.times}')"
]
},
{
"cell_type": "markdown",
"id": "afdf47c2",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"\n",
"\n"
]
},
{
"cell_type": "markdown",
"id": "9171b1f5",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Data Classes\n",
"- `AnalogSignal`: Continuous data sampled in **regular** intervals\n",
"- `IrregularlySampledSignal`: Continuous data sampled in **irregular** intervals\n",
"- `ImageSequence`: Continuous 2D **frames** sampled in regular intervals"
]
},
{
"cell_type": "markdown",
"id": "cec7e8c0",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
""
]
},
{
"cell_type": "markdown",
"id": "0bfb4ac8",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Data Classes\n",
"- `AnalogSignal`: Continuous data sampled in **regular** intervals\n",
"- `IrregularlySampledSignal`: Continuous data sampled in **irregular** intervals\n",
"- `ImageSequence`: Continuous 2D **image frames** sampled in regular intervals\n",
"- `SpikeTrain`: Time point data (& optional waveform snippet)"
]
},
{
"cell_type": "markdown",
"id": "0534eb6b",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"source": [
"*essential metadata* time values, physical units of times, (& waveform sampling rate, waveform offset to corresponding time value)"
]
},
{
"cell_type": "code",
"execution_count": 29,
"id": "d7efe06e",
"metadata": {},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"spiketrain: [1. 4. 5.7] ms\n",
"t_start & t_stop: (array(0.) * ms, array(300.) * ms)\n"
]
}
],
"source": [
"st = neo.SpikeTrain([1, 4, 5.7], units='ms', name='#001', t_start=0*pq.ms, t_stop=300*pq.ms)\n",
"print(f'spiketrain: {st}')\n",
"print(f't_start & t_stop: {st.t_start, st.t_stop}')"
]
},
{
"cell_type": "markdown",
"id": "d5ad23e7",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
""
]
},
{
"cell_type": "markdown",
"id": "d0456d48",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Data Classes\n",
"- `AnalogSignal`: Continuous data sampled in **regular** intervals\n",
"- `IrregularlySampledSignal`: Continuous data sampled in **irregular** intervals\n",
"- `ImageSequence`: Continuous 2D **image frames** sampled in regular intervals\n",
"- `SpikeTrain`: Time point data (& optional waveform snippet)\n",
"- `Event`: Experiment reference time points (e.g. trigger, trial start, ...)\n",
"- `Epoch`: Experiment reference time ranges (e.g. trial, stimulation, ...)"
]
},
{
"cell_type": "markdown",
"id": "759eee37",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
""
]
},
{
"cell_type": "markdown",
"id": "b1d5a21d",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Additional metadata attributes\n",
"- human readable label of objects via `name` attribute\n",
"- custom metadata annotations via `annotation` and `array_annotation` attributes\n",
"- `Event` and `Epoch` can be used to `label` each time point / time period"
]
},
{
"cell_type": "code",
"execution_count": 42,
"id": "321dc012",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"name: electrode 1A\n",
"annotations: {'signal_quality': 'good'}\n",
"number of channels: 2\n",
"array_annotations: {'channel_id': array([1, 2])}\n"
]
}
],
"source": [
"anasig.name = 'electrode 1A'\n",
"anasig.annotate(signal_quality='good')\n",
"anasig.array_annotate(channel_id=[1,2])\n",
"print(f'name: {anasig.name}')\n",
"print(f'annotations: {anasig.annotations}')\n",
"print(f'number of channels: {anasig.shape[-1]}')\n",
"print(f'array_annotations: {anasig.array_annotations}')"
]
},
{
"cell_type": "markdown",
"id": "c2062418",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Neo objects provide utility functions\n",
"Some usefull utility attributes and methods of neo data objects are\n",
"- `.times` to get array of corresponding time values\n",
"- `.time_slice()` to crop to a specific time range\n",
"- `.merge()` to combine multiple objects of the same type\n",
"- `.concatenate()` to append multiple signal objects\n",
"- `.downsample()` to create a new signal with a different sampling rate\n",
"- `.magnitude` to extract the underlying numpy array\n",
"- check out the [documentation](https://neo.readthedocs.io/en/latest/core.html) to discover more!"
]
},
{
"cell_type": "markdown",
"id": "0b7a117b",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"## Relations between data objects\n",
"- `ChannelView`: select a subset of channels of a signal, e.g. all even channels of an `AnalogSignal`\n",
"- **`Segment`**: contains data objects with a shared clock, e.g. a trial\n",
"- `Group`: groups data objects logically (no common clock required, e.g. `SpikeTrain`s of a neuronal unit)\n",
"- **`Block`**: contains all objects of a recording"
]
},
{
"cell_type": "markdown",
"id": "635213d6",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
""
]
},
{
"cell_type": "markdown",
"id": "63624385",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Neo structure\n",
"\n",
"```\n",
"Block 0\n",
" .segments\n",
" Segment 0\n",
" .analogsignals\n",
" AnalogSignal 0\n",
" AnalogSignal 1\n",
" .spiketrains\n",
" SpikeTrain 0\n",
" SpikeTrain 1\n",
" SpikeTrain 2\n",
" Segment 1\n",
" .analogsignals\n",
" AnalogSignal 0\n",
" AnalogSignal 1\n",
" .spiketrains\n",
" SpikeTrain 0\n",
" SpikeTrain 1\n",
" SpikeTrain 2\n",
"Block 1\n",
" .segments\n",
" Segment 0\n",
" ...\n",
"```"
]
},
{
"cell_type": "markdown",
"id": "1130f965",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Neo Class Overview\n",
""
]
},
{
"cell_type": "markdown",
"id": "d46c3064",
"metadata": {
"slideshow": {
"slide_type": "slide"
}
},
"source": [
"# Automatic generation of Neo objects\n",
""
]
},
{
"cell_type": "markdown",
"id": "9afd13ea",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Loading a recording session\n",
"- generation of a complete neo structure requires only **2 lines of code** and the name of the original recording system."
]
},
{
"cell_type": "code",
"execution_count": 51,
"id": "27a022ab",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"data": {
"text/plain": [
"Block with 1 segments, 1 groups\n",
"annotations: {'openephys_version': '0.4'}\n",
"file_origin: './neo_material/example_data/OpenEphys_SampleData_1'\n",
"# segments (N=1)\n",
"0: Segment with 1 analogsignals, 1 events, 1 spiketrains\n",
" annotations: {'openephys_version': '0.4',\n",
" 'date_created': \"'3-Oct-2018 131650'\",\n",
" 'openephys_segment_index': 1}\n",
" # analogsignals (N=1)\n",
" 0: AnalogSignal with 2 channels of length 423936; units V; datatype float32 \n",
" name: 'Signals CH'\n",
" annotations: {'stream_id': 'CH'}\n",
" sampling rate: 40000.0 Hz\n",
" time: 1.2992 s to 11.8976 s"
]
},
"execution_count": 51,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"recording_folder = './neo_material/example_data/OpenEphys_SampleData_1'\n",
"io = neo.io.OpenEphysIO(recording_folder)\n",
"block = io.read_block()\n",
"block"
]
},
{
"cell_type": "markdown",
"id": "82d2f42a",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Accessing data objects\n",
"- child objects can be accessed the corresponding attribute based on the class name (plural!). E.g. `.segments`, `.analogsignals`, `.spiketrains`\n",
"- child objects are stored in *lists*"
]
},
{
"cell_type": "code",
"execution_count": 60,
"id": "da4d5648",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"data": {
"text/plain": [
"AnalogSignal with 2 channels of length 423936; units V; datatype float32 \n",
"name: 'Signals CH'\n",
"annotations: {'stream_id': 'CH'}\n",
"sampling rate: 40000.0 Hz\n",
"time: 1.2992 s to 11.8976 s"
]
},
"execution_count": 60,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"# accessing the list of Segments\n",
"block.segments\n",
"# accessing the first AnalogSignal of a single Segment\n",
"segment = block.segments[0]\n",
"segment.analogsignals[0]"
]
},
{
"cell_type": "markdown",
"id": "d8faf977",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"Inspecting a spiketrain object"
]
},
{
"cell_type": "code",
"execution_count": 67,
"id": "18359d77",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"data": {
"text/plain": [
"SpikeTrain name: 'STp106.0n0#0' annotations: {'id': 'STp106.0n0#0'}"
]
},
"execution_count": 67,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"spiketrain = segment.spiketrains[0]\n",
"spiketrain"
]
},
{
"cell_type": "code",
"execution_count": 70,
"id": "3eed0a74",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"Number of spikes: 454\n"
]
},
{
"data": {
"text/plain": [
"array([1.30545 , 1.403875, 1.415325, 1.422425, 1.428775, 1.45465 ,\n",
" 1.4658 , 1.5147 , 1.51905 , 1.521725, 1.546 , 1.563975,\n",
" 1.566525, 1.568325, 1.573275, 1.58005 , 1.588125, 1.59505 ,\n",
" 1.599125, 1.654425]) * s"
]
},
"execution_count": 70,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"print(f'Number of spikes: {len(spiketrain)}')\n",
"spiketrain.times[:20]"
]
},
{
"cell_type": "markdown",
"id": "b338dc98",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### Dealing with large datasets\n",
"How to load only required data\n",
"\n",
"- some IOs are based on `RawIO` concept for efficient reading of data\n",
"- `RawIO`s require additional symmetries in the dataset for efficient loading\n",
"- *lazy* data objects (ProxyObjects) can be loaded using `io.read_block(`**`lazy=True`**`)`\n",
"- ProxyObjects provide a `.load(t_start, t_stop)` method that loads requested data in memory and returns complete neo data object."
]
},
{
"cell_type": "code",
"execution_count": 109,
"id": "e516de90",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"data": {
"text/plain": [
"AnalogSignalProxy name: 'Signals CH' annotations: {'stream_id': 'CH'}"
]
},
"execution_count": 109,
"metadata": {},
"output_type": "execute_result"
}
],
"source": [
"recording_folder = './neo_material/example_data/OpenEphys_SampleData_1'\n",
"io = neo.io.OpenEphysIO(recording_folder)\n",
"block = io.read_block(lazy=True)\n",
"lazy_anasig = block.segments[0].analogsignals[0]\n",
"lazy_anasig"
]
},
{
"cell_type": "markdown",
"id": "9a6a885a",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"#### Loading data from a Proxy object\n",
"- ProxyObjects contain metadata and shape information\n"
]
},
{
"cell_type": "code",
"execution_count": 90,
"id": "f50bc7a3",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"signal shape: (423936, 2)\n",
"signal sampling rate: 40000.0 Hz\n",
"signal annotations: {'stream_id': 'CH'}\n"
]
}
],
"source": [
"print(f'signal shape: {lazy_anasig.shape}')\n",
"print(f'signal sampling rate: {lazy_anasig.sampling_rate}')\n",
"print(f'signal annotations: {lazy_anasig.annotations}')"
]
},
{
"cell_type": "markdown",
"id": "e4ef7c39",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"source": [
"- data of a specific channel and time range can be loaded selectively into a new neo object"
]
},
{
"cell_type": "code",
"execution_count": 85,
"id": "2aa4d7e9",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [],
"source": [
"anasig = lazy_anasig.load(time_slice=(5*pq.s,6*pq.s), channel_indexes=[0])"
]
},
{
"cell_type": "code",
"execution_count": 93,
"id": "7a4dca6d",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"signal shape: (40000, 1)\n",
"signal sampling rate: 40000.0 Hz\n",
"signal annotations: [5. 5.000025 5.00005 ... 5.999925 5.99995 5.999975] s\n",
"signal values: [[13.45]\n",
" [13.1 ]\n",
" [10.8 ]\n",
" ...\n",
" [12.3 ]\n",
" [13.05]\n",
" [14.15]]\n"
]
}
],
"source": [
"print(f'signal shape: {anasig.shape}')\n",
"print(f'signal sampling rate: {anasig.sampling_rate}')\n",
"print(f'signal annotations: {anasig.times}')\n",
"print(f'signal values: {anasig.magnitude}')"
]
},
{
"cell_type": "markdown",
"id": "1f2d38e9",
"metadata": {
"slideshow": {
"slide_type": "subslide"
}
},
"source": [
"### How to save data?"
]
},
{
"cell_type": "markdown",
"id": "32c21898",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"source": [
"- selected open formats are supported for writing\n",
" - **NIX**[1](#fn1)\n",
" - NWB[1](#fn1)\n",
" - Matlab[2](#fn2)\n",
" - Ascii[2](#fn2)\n",
" - Numpy Pickle[3](#fn3)\n",
" \n",
"1 Support of neo-compatible format aspects\n",
"\n",
"2 Does not capture complete set of metadata\n",
"\n",
"3 Strong dependency on Numpy and Neo version"
]
},
{
"cell_type": "code",
"execution_count": 119,
"id": "18593599",
"metadata": {
"slideshow": {
"slide_type": "fragment"
}
},
"outputs": [
{
"name": "stdout",
"output_type": "stream",
"text": [
"[
"
]
}
],
"metadata": {
"celltoolbar": "Slideshow",
"kernelspec": {
"display_name": "Python 3 (ipykernel)",
"language": "python",
"name": "python3"
},
"language_info": {
"codemirror_mode": {
"name": "ipython",
"version": 3
},
"file_extension": ".py",
"mimetype": "text/x-python",
"name": "python",
"nbconvert_exporter": "python",
"pygments_lexer": "ipython3",
"version": "3.9.6"
}
},
"livereveal":{
"autolaunch": true,
"scroll": true}
},
"nbformat": 4,
"nbformat_minor": 5
}
\n",
" \n",
"
\n",
"